
Pelagibacterium luteolum
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Devosiaceae; Pelagibacterium
Average proteome isoelectric point is 6.22
Get precalculated fractions of proteins
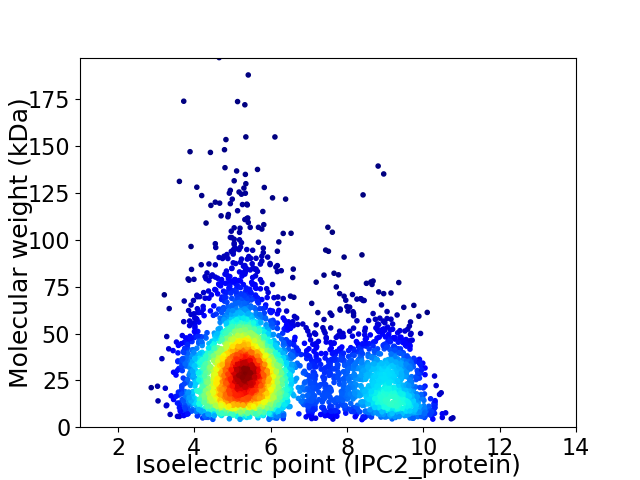
Virtual 2D-PAGE plot for 4253 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1G7S4M6|A0A1G7S4M6_9RHIZ Predicted aconitase subunit 1 OS=Pelagibacterium luteolum OX=440168 GN=SAMN04487974_101300 PE=4 SV=1
MM1 pKa = 7.39QEE3 pKa = 3.71FEE5 pKa = 4.26TTFRR9 pKa = 11.84QGVNGYY15 pKa = 10.42ASTVDD20 pKa = 3.64TQLLQANTGANHH32 pKa = 7.09ANTAVLGVDD41 pKa = 3.62RR42 pKa = 11.84NSHH45 pKa = 4.4VQVLLRR51 pKa = 11.84FDD53 pKa = 5.03DD54 pKa = 4.14LFGNAVSQVPAGAEE68 pKa = 3.82ILSATLTLHH77 pKa = 5.2TTNTGDD83 pKa = 3.23GATLHH88 pKa = 6.93RR89 pKa = 11.84MLTNWSGNSTWSSTGSGIQTNGVEE113 pKa = 4.32ALAAADD119 pKa = 3.9VNTGSTNLGASNFDD133 pKa = 3.24VTASVQAWAGGQANLGWVFVSLGTNGWDD161 pKa = 3.75FYY163 pKa = 10.78SAQGATPPQLTIRR176 pKa = 11.84YY177 pKa = 8.8RR178 pKa = 11.84IEE180 pKa = 4.37GEE182 pKa = 4.0PNTAPVANPDD192 pKa = 3.74TVVVDD197 pKa = 3.87EE198 pKa = 5.55DD199 pKa = 3.97GSATVNVLANDD210 pKa = 3.67ADD212 pKa = 4.26ADD214 pKa = 3.95GDD216 pKa = 4.17TLVVSGVGTPANGVAVLNGNGTITYY241 pKa = 7.59TPHH244 pKa = 7.33ANFFGNDD251 pKa = 3.22SFSYY255 pKa = 9.82TLSDD259 pKa = 3.41GKK261 pKa = 10.92GGQSTGQVSVTVNPVNDD278 pKa = 3.79APVATGDD285 pKa = 3.61TASVNEE291 pKa = 4.28GGNVVVTVLSNDD303 pKa = 3.08SDD305 pKa = 3.75IDD307 pKa = 3.68GDD309 pKa = 4.05ALSVSGVGTPSNGTAVINSNGTITYY334 pKa = 7.68TPNAGYY340 pKa = 8.33TGPDD344 pKa = 3.12SFSYY348 pKa = 10.07TINDD352 pKa = 3.8GKK354 pKa = 11.09GGTATASVAVTVHH367 pKa = 6.42PVNDD371 pKa = 3.74APVATGDD378 pKa = 3.75SVSVNEE384 pKa = 4.87DD385 pKa = 3.15GAVAIAVLANDD396 pKa = 4.48SDD398 pKa = 4.43PDD400 pKa = 4.13GDD402 pKa = 3.94VLTVTGVGTPAHH414 pKa = 6.41GLAIINSNGTITYY427 pKa = 7.75TPNANYY433 pKa = 9.95HH434 pKa = 6.08GPDD437 pKa = 3.08SFTYY441 pKa = 9.85TISDD445 pKa = 3.59GKK447 pKa = 10.7GGQATASVSVTVNSVNDD464 pKa = 3.68VPVADD469 pKa = 4.74ADD471 pKa = 3.76AASVIEE477 pKa = 4.51GGSVTVNVLSNDD489 pKa = 3.05SDD491 pKa = 4.31ADD493 pKa = 3.57GDD495 pKa = 4.13ALSVIGVGTPANGNAVLNGNGTITYY520 pKa = 7.65TPNAGYY526 pKa = 8.35TGPDD530 pKa = 3.2GFSYY534 pKa = 10.36TISDD538 pKa = 3.72GKK540 pKa = 10.82GGQATANVAITVNSDD555 pKa = 3.34TPNTVPVATSDD566 pKa = 3.73DD567 pKa = 3.72TSVLVDD573 pKa = 3.47GSVTVKK579 pKa = 10.51VLANDD584 pKa = 3.44YY585 pKa = 11.32DD586 pKa = 4.72LDD588 pKa = 4.84GDD590 pKa = 4.09SLAIISVSGPASGFAVVNPNGTITYY615 pKa = 7.62TPNAGYY621 pKa = 10.16SGPDD625 pKa = 2.93SFTYY629 pKa = 9.81TVADD633 pKa = 4.11GKK635 pKa = 11.24GGTAVGSVNVNVVEE649 pKa = 4.21PVSGDD654 pKa = 3.37VEE656 pKa = 4.33HH657 pKa = 6.77QVTFRR662 pKa = 11.84QGVNGYY668 pKa = 9.77SGTVDD673 pKa = 3.45TQILQSNTGANHH685 pKa = 6.95ANTAVIGVDD694 pKa = 2.78RR695 pKa = 11.84GTDD698 pKa = 3.41VQVLLRR704 pKa = 11.84FNDD707 pKa = 3.55LFGNGAFQVPPGSEE721 pKa = 3.71ILSATLTLQTTNTGNGATLHH741 pKa = 6.73RR742 pKa = 11.84MLTNWSDD749 pKa = 3.05TSTWASIVSGVQTNGVEE766 pKa = 4.14ALTAADD772 pKa = 3.64VRR774 pKa = 11.84TGATNLGASSYY785 pKa = 11.31DD786 pKa = 3.26VTASVQAWAGGQANLGWVFVSLGTNGWDD814 pKa = 3.77FYY816 pKa = 11.0SAQGSIPPQLTITYY830 pKa = 7.64RR831 pKa = 11.84APGGQEE837 pKa = 3.54NVAPTAAIDD846 pKa = 3.57AGVVNEE852 pKa = 5.01DD853 pKa = 3.25GSVVINPLANDD864 pKa = 3.79TDD866 pKa = 4.53PDD868 pKa = 4.09GGTLTIVGIGTPTNGVAVLNSNGTVTYY895 pKa = 9.58TPNANYY901 pKa = 10.19NGPDD905 pKa = 3.3SFSYY909 pKa = 10.21TISDD913 pKa = 3.94GQGGTASGTITIVVKK928 pKa = 9.72PVNDD932 pKa = 3.69APVAAGDD939 pKa = 4.03GAVVTQGGLVVVDD952 pKa = 4.32VLANDD957 pKa = 3.74SDD959 pKa = 4.12VDD961 pKa = 3.77GDD963 pKa = 4.05ALSVIGVGTPAHH975 pKa = 7.05GIAVINSDD983 pKa = 3.37GTVTYY988 pKa = 9.28TPAGGYY994 pKa = 9.52SGSDD998 pKa = 3.07SFSYY1002 pKa = 10.22TISDD1006 pKa = 3.67GKK1008 pKa = 10.88GGEE1011 pKa = 4.32ATGTAAVTVNASVPVNTPPVANPDD1035 pKa = 4.07SISVNEE1041 pKa = 4.79DD1042 pKa = 2.78GSVVISVLANDD1053 pKa = 3.84TDD1055 pKa = 4.66ANGHH1059 pKa = 6.3ALTVTGVGTPGHH1071 pKa = 5.59GTVVINGNGTITYY1084 pKa = 7.9TPTANYY1090 pKa = 9.12HH1091 pKa = 5.99GPDD1094 pKa = 3.24SFTYY1098 pKa = 9.83TISDD1102 pKa = 3.53GHH1104 pKa = 6.91GGAATGAVSITVNPVNDD1121 pKa = 3.84APVANADD1128 pKa = 4.08TIAVNEE1134 pKa = 4.24DD1135 pKa = 3.11AAVVISVLANDD1146 pKa = 3.95TDD1148 pKa = 3.8IDD1150 pKa = 4.48GDD1152 pKa = 4.03TLTVIGVGAPAHH1164 pKa = 6.16GSAVVNSNGTITYY1177 pKa = 7.89TPSTNYY1183 pKa = 10.39NGADD1187 pKa = 3.12SFTYY1191 pKa = 9.9TISDD1195 pKa = 3.59GKK1197 pKa = 10.7GGQATGTVNVTVNPINDD1214 pKa = 3.65APVANADD1221 pKa = 3.65SAFVQVGASVVVPVLLNDD1239 pKa = 3.24VDD1241 pKa = 3.93VDD1243 pKa = 3.86GDD1245 pKa = 3.95ALTVTGFAVAPQHH1258 pKa = 5.3GTAVVNGNGTVTYY1271 pKa = 8.89TPSAGYY1277 pKa = 10.35VGVDD1281 pKa = 2.79AFIYY1285 pKa = 9.28TISDD1289 pKa = 3.45GKK1291 pKa = 10.79GGSASATVTVNVTEE1305 pKa = 4.33QPDD1308 pKa = 3.99DD1309 pKa = 4.19PLHH1312 pKa = 6.49ASYY1315 pKa = 10.66IATAISGSTSARR1327 pKa = 11.84HH1328 pKa = 5.96LEE1330 pKa = 4.13HH1331 pKa = 7.15SNSTKK1336 pKa = 10.44SFYY1339 pKa = 10.63FAGAWWAILPNSANWSIHH1357 pKa = 5.35KK1358 pKa = 10.58FNGTTPDD1365 pKa = 3.36EE1366 pKa = 4.55GQLGGWTIKK1375 pKa = 9.47STALFASNSSNRR1387 pKa = 11.84ADD1389 pKa = 4.15LAWDD1393 pKa = 3.53PVTEE1397 pKa = 4.13TLYY1400 pKa = 11.37VMQYY1404 pKa = 11.12NNGTPRR1410 pKa = 11.84LFALDD1415 pKa = 3.72YY1416 pKa = 11.25AAGTDD1421 pKa = 3.33SWVVGSNIVLAGSGGVLSGSQWTSNIEE1448 pKa = 3.69MRR1450 pKa = 11.84LSIDD1454 pKa = 3.38QNGVPVVSAIGASGSSAGLHH1474 pKa = 6.26LGFATSSNLSTWGQVLLDD1492 pKa = 3.69ATTDD1496 pKa = 3.28RR1497 pKa = 11.84SGGSNGDD1504 pKa = 3.42SKK1506 pKa = 11.93ADD1508 pKa = 2.98IVFFTQGGANKK1519 pKa = 9.86IAVIYY1524 pKa = 10.56SRR1526 pKa = 11.84DD1527 pKa = 3.52GAGSDD1532 pKa = 3.93NDD1534 pKa = 3.62WSIVWRR1540 pKa = 11.84DD1541 pKa = 3.86TPQSAADD1548 pKa = 4.23YY1549 pKa = 10.08VNGWQKK1555 pKa = 10.75EE1556 pKa = 4.4VITDD1560 pKa = 3.52DD1561 pKa = 4.03VNIDD1565 pKa = 2.91NHH1567 pKa = 5.6MAAVSDD1573 pKa = 3.63GTTIYY1578 pKa = 11.12AVIKK1582 pKa = 10.83DD1583 pKa = 4.41DD1584 pKa = 6.67DD1585 pKa = 4.05NSLWLLKK1592 pKa = 9.82GHH1594 pKa = 7.08PGNWDD1599 pKa = 3.28APLLIVEE1606 pKa = 5.49GGAHH1610 pKa = 6.63DD1611 pKa = 4.07PSRR1614 pKa = 11.84PGIVLDD1620 pKa = 3.73HH1621 pKa = 6.33TNDD1624 pKa = 3.29RR1625 pKa = 11.84LFIYY1629 pKa = 9.62YY1630 pKa = 9.53QEE1632 pKa = 4.35STSDD1636 pKa = 3.42PEE1638 pKa = 4.12GAVFLKK1644 pKa = 9.94VTDD1647 pKa = 4.1ADD1649 pKa = 3.85NPTFDD1654 pKa = 4.23SDD1656 pKa = 4.87DD1657 pKa = 3.45IGMMILQGTNASHH1670 pKa = 7.81DD1671 pKa = 3.69MLDD1674 pKa = 3.74PQRR1677 pKa = 11.84PAHH1680 pKa = 6.78AVGTDD1685 pKa = 3.06TGNEE1689 pKa = 4.14FIILAKK1695 pKa = 10.2NQQTNTIWYY1704 pKa = 9.12NDD1706 pKa = 2.96VDD1708 pKa = 4.5LGFEE1712 pKa = 3.99YY1713 pKa = 10.68SIAA1716 pKa = 3.67
MM1 pKa = 7.39QEE3 pKa = 3.71FEE5 pKa = 4.26TTFRR9 pKa = 11.84QGVNGYY15 pKa = 10.42ASTVDD20 pKa = 3.64TQLLQANTGANHH32 pKa = 7.09ANTAVLGVDD41 pKa = 3.62RR42 pKa = 11.84NSHH45 pKa = 4.4VQVLLRR51 pKa = 11.84FDD53 pKa = 5.03DD54 pKa = 4.14LFGNAVSQVPAGAEE68 pKa = 3.82ILSATLTLHH77 pKa = 5.2TTNTGDD83 pKa = 3.23GATLHH88 pKa = 6.93RR89 pKa = 11.84MLTNWSGNSTWSSTGSGIQTNGVEE113 pKa = 4.32ALAAADD119 pKa = 3.9VNTGSTNLGASNFDD133 pKa = 3.24VTASVQAWAGGQANLGWVFVSLGTNGWDD161 pKa = 3.75FYY163 pKa = 10.78SAQGATPPQLTIRR176 pKa = 11.84YY177 pKa = 8.8RR178 pKa = 11.84IEE180 pKa = 4.37GEE182 pKa = 4.0PNTAPVANPDD192 pKa = 3.74TVVVDD197 pKa = 3.87EE198 pKa = 5.55DD199 pKa = 3.97GSATVNVLANDD210 pKa = 3.67ADD212 pKa = 4.26ADD214 pKa = 3.95GDD216 pKa = 4.17TLVVSGVGTPANGVAVLNGNGTITYY241 pKa = 7.59TPHH244 pKa = 7.33ANFFGNDD251 pKa = 3.22SFSYY255 pKa = 9.82TLSDD259 pKa = 3.41GKK261 pKa = 10.92GGQSTGQVSVTVNPVNDD278 pKa = 3.79APVATGDD285 pKa = 3.61TASVNEE291 pKa = 4.28GGNVVVTVLSNDD303 pKa = 3.08SDD305 pKa = 3.75IDD307 pKa = 3.68GDD309 pKa = 4.05ALSVSGVGTPSNGTAVINSNGTITYY334 pKa = 7.68TPNAGYY340 pKa = 8.33TGPDD344 pKa = 3.12SFSYY348 pKa = 10.07TINDD352 pKa = 3.8GKK354 pKa = 11.09GGTATASVAVTVHH367 pKa = 6.42PVNDD371 pKa = 3.74APVATGDD378 pKa = 3.75SVSVNEE384 pKa = 4.87DD385 pKa = 3.15GAVAIAVLANDD396 pKa = 4.48SDD398 pKa = 4.43PDD400 pKa = 4.13GDD402 pKa = 3.94VLTVTGVGTPAHH414 pKa = 6.41GLAIINSNGTITYY427 pKa = 7.75TPNANYY433 pKa = 9.95HH434 pKa = 6.08GPDD437 pKa = 3.08SFTYY441 pKa = 9.85TISDD445 pKa = 3.59GKK447 pKa = 10.7GGQATASVSVTVNSVNDD464 pKa = 3.68VPVADD469 pKa = 4.74ADD471 pKa = 3.76AASVIEE477 pKa = 4.51GGSVTVNVLSNDD489 pKa = 3.05SDD491 pKa = 4.31ADD493 pKa = 3.57GDD495 pKa = 4.13ALSVIGVGTPANGNAVLNGNGTITYY520 pKa = 7.65TPNAGYY526 pKa = 8.35TGPDD530 pKa = 3.2GFSYY534 pKa = 10.36TISDD538 pKa = 3.72GKK540 pKa = 10.82GGQATANVAITVNSDD555 pKa = 3.34TPNTVPVATSDD566 pKa = 3.73DD567 pKa = 3.72TSVLVDD573 pKa = 3.47GSVTVKK579 pKa = 10.51VLANDD584 pKa = 3.44YY585 pKa = 11.32DD586 pKa = 4.72LDD588 pKa = 4.84GDD590 pKa = 4.09SLAIISVSGPASGFAVVNPNGTITYY615 pKa = 7.62TPNAGYY621 pKa = 10.16SGPDD625 pKa = 2.93SFTYY629 pKa = 9.81TVADD633 pKa = 4.11GKK635 pKa = 11.24GGTAVGSVNVNVVEE649 pKa = 4.21PVSGDD654 pKa = 3.37VEE656 pKa = 4.33HH657 pKa = 6.77QVTFRR662 pKa = 11.84QGVNGYY668 pKa = 9.77SGTVDD673 pKa = 3.45TQILQSNTGANHH685 pKa = 6.95ANTAVIGVDD694 pKa = 2.78RR695 pKa = 11.84GTDD698 pKa = 3.41VQVLLRR704 pKa = 11.84FNDD707 pKa = 3.55LFGNGAFQVPPGSEE721 pKa = 3.71ILSATLTLQTTNTGNGATLHH741 pKa = 6.73RR742 pKa = 11.84MLTNWSDD749 pKa = 3.05TSTWASIVSGVQTNGVEE766 pKa = 4.14ALTAADD772 pKa = 3.64VRR774 pKa = 11.84TGATNLGASSYY785 pKa = 11.31DD786 pKa = 3.26VTASVQAWAGGQANLGWVFVSLGTNGWDD814 pKa = 3.77FYY816 pKa = 11.0SAQGSIPPQLTITYY830 pKa = 7.64RR831 pKa = 11.84APGGQEE837 pKa = 3.54NVAPTAAIDD846 pKa = 3.57AGVVNEE852 pKa = 5.01DD853 pKa = 3.25GSVVINPLANDD864 pKa = 3.79TDD866 pKa = 4.53PDD868 pKa = 4.09GGTLTIVGIGTPTNGVAVLNSNGTVTYY895 pKa = 9.58TPNANYY901 pKa = 10.19NGPDD905 pKa = 3.3SFSYY909 pKa = 10.21TISDD913 pKa = 3.94GQGGTASGTITIVVKK928 pKa = 9.72PVNDD932 pKa = 3.69APVAAGDD939 pKa = 4.03GAVVTQGGLVVVDD952 pKa = 4.32VLANDD957 pKa = 3.74SDD959 pKa = 4.12VDD961 pKa = 3.77GDD963 pKa = 4.05ALSVIGVGTPAHH975 pKa = 7.05GIAVINSDD983 pKa = 3.37GTVTYY988 pKa = 9.28TPAGGYY994 pKa = 9.52SGSDD998 pKa = 3.07SFSYY1002 pKa = 10.22TISDD1006 pKa = 3.67GKK1008 pKa = 10.88GGEE1011 pKa = 4.32ATGTAAVTVNASVPVNTPPVANPDD1035 pKa = 4.07SISVNEE1041 pKa = 4.79DD1042 pKa = 2.78GSVVISVLANDD1053 pKa = 3.84TDD1055 pKa = 4.66ANGHH1059 pKa = 6.3ALTVTGVGTPGHH1071 pKa = 5.59GTVVINGNGTITYY1084 pKa = 7.9TPTANYY1090 pKa = 9.12HH1091 pKa = 5.99GPDD1094 pKa = 3.24SFTYY1098 pKa = 9.83TISDD1102 pKa = 3.53GHH1104 pKa = 6.91GGAATGAVSITVNPVNDD1121 pKa = 3.84APVANADD1128 pKa = 4.08TIAVNEE1134 pKa = 4.24DD1135 pKa = 3.11AAVVISVLANDD1146 pKa = 3.95TDD1148 pKa = 3.8IDD1150 pKa = 4.48GDD1152 pKa = 4.03TLTVIGVGAPAHH1164 pKa = 6.16GSAVVNSNGTITYY1177 pKa = 7.89TPSTNYY1183 pKa = 10.39NGADD1187 pKa = 3.12SFTYY1191 pKa = 9.9TISDD1195 pKa = 3.59GKK1197 pKa = 10.7GGQATGTVNVTVNPINDD1214 pKa = 3.65APVANADD1221 pKa = 3.65SAFVQVGASVVVPVLLNDD1239 pKa = 3.24VDD1241 pKa = 3.93VDD1243 pKa = 3.86GDD1245 pKa = 3.95ALTVTGFAVAPQHH1258 pKa = 5.3GTAVVNGNGTVTYY1271 pKa = 8.89TPSAGYY1277 pKa = 10.35VGVDD1281 pKa = 2.79AFIYY1285 pKa = 9.28TISDD1289 pKa = 3.45GKK1291 pKa = 10.79GGSASATVTVNVTEE1305 pKa = 4.33QPDD1308 pKa = 3.99DD1309 pKa = 4.19PLHH1312 pKa = 6.49ASYY1315 pKa = 10.66IATAISGSTSARR1327 pKa = 11.84HH1328 pKa = 5.96LEE1330 pKa = 4.13HH1331 pKa = 7.15SNSTKK1336 pKa = 10.44SFYY1339 pKa = 10.63FAGAWWAILPNSANWSIHH1357 pKa = 5.35KK1358 pKa = 10.58FNGTTPDD1365 pKa = 3.36EE1366 pKa = 4.55GQLGGWTIKK1375 pKa = 9.47STALFASNSSNRR1387 pKa = 11.84ADD1389 pKa = 4.15LAWDD1393 pKa = 3.53PVTEE1397 pKa = 4.13TLYY1400 pKa = 11.37VMQYY1404 pKa = 11.12NNGTPRR1410 pKa = 11.84LFALDD1415 pKa = 3.72YY1416 pKa = 11.25AAGTDD1421 pKa = 3.33SWVVGSNIVLAGSGGVLSGSQWTSNIEE1448 pKa = 3.69MRR1450 pKa = 11.84LSIDD1454 pKa = 3.38QNGVPVVSAIGASGSSAGLHH1474 pKa = 6.26LGFATSSNLSTWGQVLLDD1492 pKa = 3.69ATTDD1496 pKa = 3.28RR1497 pKa = 11.84SGGSNGDD1504 pKa = 3.42SKK1506 pKa = 11.93ADD1508 pKa = 2.98IVFFTQGGANKK1519 pKa = 9.86IAVIYY1524 pKa = 10.56SRR1526 pKa = 11.84DD1527 pKa = 3.52GAGSDD1532 pKa = 3.93NDD1534 pKa = 3.62WSIVWRR1540 pKa = 11.84DD1541 pKa = 3.86TPQSAADD1548 pKa = 4.23YY1549 pKa = 10.08VNGWQKK1555 pKa = 10.75EE1556 pKa = 4.4VITDD1560 pKa = 3.52DD1561 pKa = 4.03VNIDD1565 pKa = 2.91NHH1567 pKa = 5.6MAAVSDD1573 pKa = 3.63GTTIYY1578 pKa = 11.12AVIKK1582 pKa = 10.83DD1583 pKa = 4.41DD1584 pKa = 6.67DD1585 pKa = 4.05NSLWLLKK1592 pKa = 9.82GHH1594 pKa = 7.08PGNWDD1599 pKa = 3.28APLLIVEE1606 pKa = 5.49GGAHH1610 pKa = 6.63DD1611 pKa = 4.07PSRR1614 pKa = 11.84PGIVLDD1620 pKa = 3.73HH1621 pKa = 6.33TNDD1624 pKa = 3.29RR1625 pKa = 11.84LFIYY1629 pKa = 9.62YY1630 pKa = 9.53QEE1632 pKa = 4.35STSDD1636 pKa = 3.42PEE1638 pKa = 4.12GAVFLKK1644 pKa = 9.94VTDD1647 pKa = 4.1ADD1649 pKa = 3.85NPTFDD1654 pKa = 4.23SDD1656 pKa = 4.87DD1657 pKa = 3.45IGMMILQGTNASHH1670 pKa = 7.81DD1671 pKa = 3.69MLDD1674 pKa = 3.74PQRR1677 pKa = 11.84PAHH1680 pKa = 6.78AVGTDD1685 pKa = 3.06TGNEE1689 pKa = 4.14FIILAKK1695 pKa = 10.2NQQTNTIWYY1704 pKa = 9.12NDD1706 pKa = 2.96VDD1708 pKa = 4.5LGFEE1712 pKa = 3.99YY1713 pKa = 10.68SIAA1716 pKa = 3.67
Molecular weight: 174.08 kDa
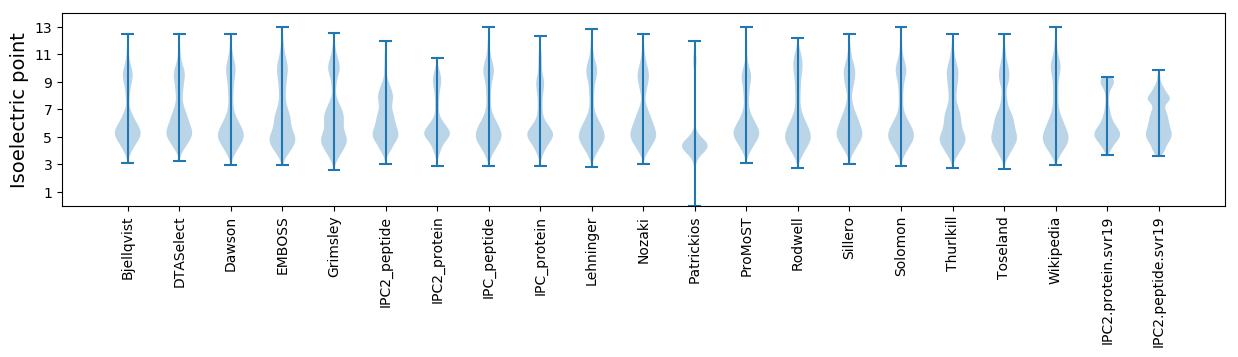
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1G7VAS4|A0A1G7VAS4_9RHIZ Antirestriction protein OS=Pelagibacterium luteolum OX=440168 GN=SAMN04487974_10423 PE=4 SV=1
MM1 pKa = 7.17SRR3 pKa = 11.84CRR5 pKa = 11.84GQSAVSGAPSLFEE18 pKa = 3.87TANTICRR25 pKa = 11.84KK26 pKa = 9.19FALSDD31 pKa = 3.32RR32 pKa = 11.84TVQRR36 pKa = 11.84MRR38 pKa = 11.84AANDD42 pKa = 3.02EE43 pKa = 4.44HH44 pKa = 9.15ALFTDD49 pKa = 3.88DD50 pKa = 5.43DD51 pKa = 4.2AANTEE56 pKa = 4.25VQLGHH61 pKa = 6.71TGGQVGVEE69 pKa = 4.47DD70 pKa = 5.15SSDD73 pKa = 3.56DD74 pKa = 4.9HH75 pKa = 8.77IGTAQKK81 pKa = 9.18NTINGPYY88 pKa = 9.91RR89 pKa = 11.84SIRR92 pKa = 11.84PLGRR96 pKa = 11.84FSTRR100 pKa = 11.84QWAWRR105 pKa = 11.84STSLKK110 pKa = 9.2TDD112 pKa = 3.73PSSCSAGRR120 pKa = 11.84ASSCTNTAAEE130 pKa = 4.24KK131 pKa = 9.26LTYY134 pKa = 10.6SRR136 pKa = 11.84VAPPLPRR143 pKa = 11.84RR144 pKa = 11.84AQAAIRR150 pKa = 11.84SAKK153 pKa = 9.73PRR155 pKa = 11.84VLRR158 pKa = 11.84TASMKK163 pKa = 10.56AFSRR167 pKa = 11.84SSRR170 pKa = 11.84SASFAMAA177 pKa = 4.46
MM1 pKa = 7.17SRR3 pKa = 11.84CRR5 pKa = 11.84GQSAVSGAPSLFEE18 pKa = 3.87TANTICRR25 pKa = 11.84KK26 pKa = 9.19FALSDD31 pKa = 3.32RR32 pKa = 11.84TVQRR36 pKa = 11.84MRR38 pKa = 11.84AANDD42 pKa = 3.02EE43 pKa = 4.44HH44 pKa = 9.15ALFTDD49 pKa = 3.88DD50 pKa = 5.43DD51 pKa = 4.2AANTEE56 pKa = 4.25VQLGHH61 pKa = 6.71TGGQVGVEE69 pKa = 4.47DD70 pKa = 5.15SSDD73 pKa = 3.56DD74 pKa = 4.9HH75 pKa = 8.77IGTAQKK81 pKa = 9.18NTINGPYY88 pKa = 9.91RR89 pKa = 11.84SIRR92 pKa = 11.84PLGRR96 pKa = 11.84FSTRR100 pKa = 11.84QWAWRR105 pKa = 11.84STSLKK110 pKa = 9.2TDD112 pKa = 3.73PSSCSAGRR120 pKa = 11.84ASSCTNTAAEE130 pKa = 4.24KK131 pKa = 9.26LTYY134 pKa = 10.6SRR136 pKa = 11.84VAPPLPRR143 pKa = 11.84RR144 pKa = 11.84AQAAIRR150 pKa = 11.84SAKK153 pKa = 9.73PRR155 pKa = 11.84VLRR158 pKa = 11.84TASMKK163 pKa = 10.56AFSRR167 pKa = 11.84SSRR170 pKa = 11.84SASFAMAA177 pKa = 4.46
Molecular weight: 19.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1271227 |
39 |
1837 |
298.9 |
32.47 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.1 ± 0.052 | 0.717 ± 0.01 |
5.996 ± 0.037 | 5.889 ± 0.041 |
3.841 ± 0.025 | 8.36 ± 0.038 |
2.022 ± 0.019 | 5.785 ± 0.029 |
3.063 ± 0.034 | 9.966 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.599 ± 0.017 | 2.751 ± 0.022 |
4.963 ± 0.03 | 3.21 ± 0.024 |
6.641 ± 0.037 | 5.597 ± 0.026 |
5.562 ± 0.026 | 7.386 ± 0.03 |
1.311 ± 0.016 | 2.24 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
