
Monoraphidium neglectum
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Chlorophyta; core chlorophytes; Chlorophyceae; CS clade; Sphaeropleales; Selenastraceae; Monoraphidium
Average proteome isoelectric point is 6.86
Get precalculated fractions of proteins
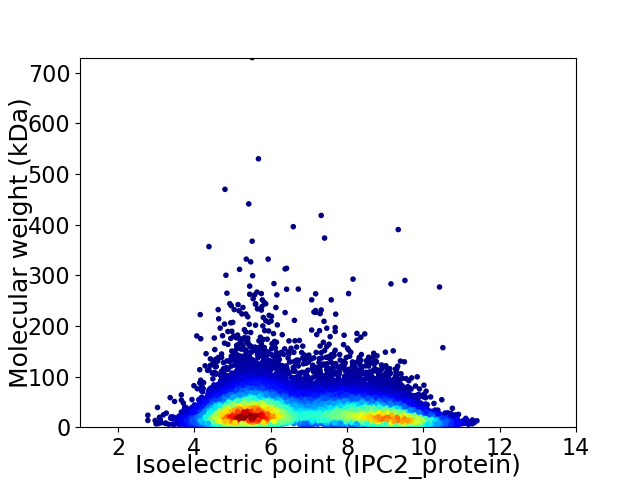
Virtual 2D-PAGE plot for 16738 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0D2MJY5|A0A0D2MJY5_9CHLO 20S proteasome subunit beta 6 OS=Monoraphidium neglectum OX=145388 GN=MNEG_4662 PE=4 SV=1
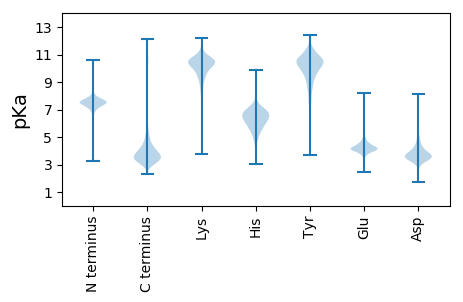
MM1 pKa = 7.72ALYY4 pKa = 10.68DD5 pKa = 4.05LFNNSGFRR13 pKa = 11.84HH14 pKa = 5.73YY15 pKa = 10.69ATGTPCFDD23 pKa = 3.04AHH25 pKa = 6.99DD26 pKa = 4.45ALRR29 pKa = 11.84DD30 pKa = 4.06PITPCEE36 pKa = 3.85ASVASGSSCFDD47 pKa = 4.27DD48 pKa = 4.45DD49 pKa = 4.73CAMSSTDD56 pKa = 3.98LFGASSSSRR65 pKa = 11.84SSADD69 pKa = 3.34SADD72 pKa = 3.92QPSLPLPRR80 pKa = 11.84SWFARR85 pKa = 11.84LSPQVPSCGIADD97 pKa = 4.05TPGGPSGGGDD107 pKa = 3.45FSSKK111 pKa = 10.51EE112 pKa = 3.47DD113 pKa = 3.18LAYY116 pKa = 10.7FNEE119 pKa = 4.08RR120 pKa = 11.84HH121 pKa = 6.07RR122 pKa = 11.84ARR124 pKa = 11.84LLSAGFSDD132 pKa = 6.53DD133 pKa = 3.85EE134 pKa = 4.46LCYY137 pKa = 10.94DD138 pKa = 5.44DD139 pKa = 6.22GGLLLGPIPFDD150 pKa = 4.05CSGDD154 pKa = 3.87DD155 pKa = 3.88GAGGAPAPRR164 pKa = 11.84PDD166 pKa = 3.84CPVVVDD172 pKa = 4.17EE173 pKa = 4.87QDD175 pKa = 3.99SISAAPGAYY184 pKa = 9.52DD185 pKa = 4.02AAFADD190 pKa = 4.81EE191 pKa = 4.29IDD193 pKa = 4.02AFFAAAGEE201 pKa = 4.51APFNPWRR208 pKa = 11.84QAPEE212 pKa = 4.04EE213 pKa = 4.25PFSFAAAAGPSGLGSVVGGGSLSFFMGVPLDD244 pKa = 4.21LEE246 pKa = 4.37IFSAPTAGGACAAVADD262 pKa = 4.22EE263 pKa = 4.3QVAISAAPGVSAGSGAKK280 pKa = 10.32GPAFDD285 pKa = 4.0FTFAGVGAEE294 pKa = 5.34GCCCWIGGDD303 pKa = 3.69VFAAASSGNGGGGSSGSSGDD323 pKa = 3.62SGSGSVGGNGGGGIGGGGGGGGDD346 pKa = 3.26PVAARR351 pKa = 11.84TRR353 pKa = 11.84SKK355 pKa = 11.19GGGAGASAAAA365 pKa = 4.19
MM1 pKa = 7.72ALYY4 pKa = 10.68DD5 pKa = 4.05LFNNSGFRR13 pKa = 11.84HH14 pKa = 5.73YY15 pKa = 10.69ATGTPCFDD23 pKa = 3.04AHH25 pKa = 6.99DD26 pKa = 4.45ALRR29 pKa = 11.84DD30 pKa = 4.06PITPCEE36 pKa = 3.85ASVASGSSCFDD47 pKa = 4.27DD48 pKa = 4.45DD49 pKa = 4.73CAMSSTDD56 pKa = 3.98LFGASSSSRR65 pKa = 11.84SSADD69 pKa = 3.34SADD72 pKa = 3.92QPSLPLPRR80 pKa = 11.84SWFARR85 pKa = 11.84LSPQVPSCGIADD97 pKa = 4.05TPGGPSGGGDD107 pKa = 3.45FSSKK111 pKa = 10.51EE112 pKa = 3.47DD113 pKa = 3.18LAYY116 pKa = 10.7FNEE119 pKa = 4.08RR120 pKa = 11.84HH121 pKa = 6.07RR122 pKa = 11.84ARR124 pKa = 11.84LLSAGFSDD132 pKa = 6.53DD133 pKa = 3.85EE134 pKa = 4.46LCYY137 pKa = 10.94DD138 pKa = 5.44DD139 pKa = 6.22GGLLLGPIPFDD150 pKa = 4.05CSGDD154 pKa = 3.87DD155 pKa = 3.88GAGGAPAPRR164 pKa = 11.84PDD166 pKa = 3.84CPVVVDD172 pKa = 4.17EE173 pKa = 4.87QDD175 pKa = 3.99SISAAPGAYY184 pKa = 9.52DD185 pKa = 4.02AAFADD190 pKa = 4.81EE191 pKa = 4.29IDD193 pKa = 4.02AFFAAAGEE201 pKa = 4.51APFNPWRR208 pKa = 11.84QAPEE212 pKa = 4.04EE213 pKa = 4.25PFSFAAAAGPSGLGSVVGGGSLSFFMGVPLDD244 pKa = 4.21LEE246 pKa = 4.37IFSAPTAGGACAAVADD262 pKa = 4.22EE263 pKa = 4.3QVAISAAPGVSAGSGAKK280 pKa = 10.32GPAFDD285 pKa = 4.0FTFAGVGAEE294 pKa = 5.34GCCCWIGGDD303 pKa = 3.69VFAAASSGNGGGGSSGSSGDD323 pKa = 3.62SGSGSVGGNGGGGIGGGGGGGGDD346 pKa = 3.26PVAARR351 pKa = 11.84TRR353 pKa = 11.84SKK355 pKa = 11.19GGGAGASAAAA365 pKa = 4.19
Molecular weight: 35.45 kDa
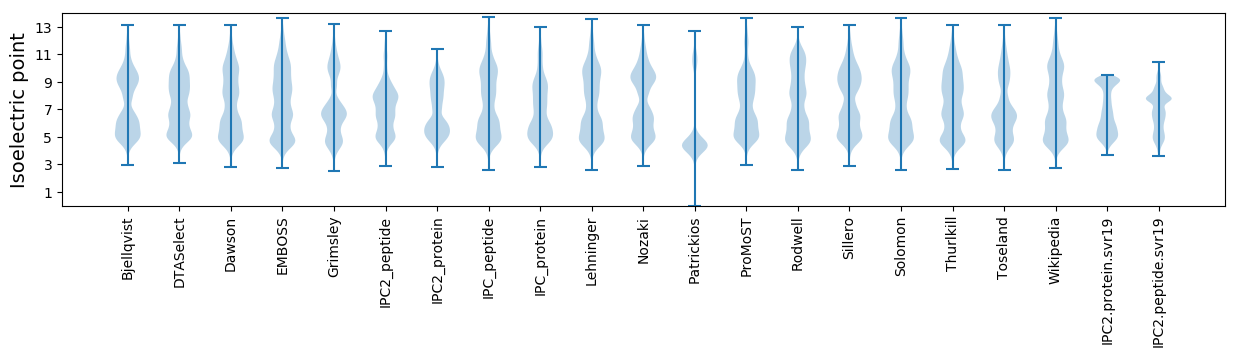
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0D2MA77|A0A0D2MA77_9CHLO AP-1 complex subunit beta-1 OS=Monoraphidium neglectum OX=145388 GN=MNEG_7799 PE=4 SV=1
AA1 pKa = 7.34QRR3 pKa = 11.84AAPGRR8 pKa = 11.84LAARR12 pKa = 11.84PRR14 pKa = 11.84GGHH17 pKa = 6.82LGRR20 pKa = 11.84PVRR23 pKa = 11.84GRR25 pKa = 11.84PDD27 pKa = 2.61WRR29 pKa = 11.84AGRR32 pKa = 11.84PRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84AGGRR41 pKa = 11.84HH42 pKa = 4.94GLAARR47 pKa = 11.84RR48 pKa = 11.84ARR50 pKa = 11.84RR51 pKa = 11.84GAGAGVARR59 pKa = 11.84ARR61 pKa = 11.84GGARR65 pKa = 11.84GGQQRR70 pKa = 11.84RR71 pKa = 11.84AGRR74 pKa = 11.84GRR76 pKa = 11.84LL77 pKa = 3.45
AA1 pKa = 7.34QRR3 pKa = 11.84AAPGRR8 pKa = 11.84LAARR12 pKa = 11.84PRR14 pKa = 11.84GGHH17 pKa = 6.82LGRR20 pKa = 11.84PVRR23 pKa = 11.84GRR25 pKa = 11.84PDD27 pKa = 2.61WRR29 pKa = 11.84AGRR32 pKa = 11.84PRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84AGGRR41 pKa = 11.84HH42 pKa = 4.94GLAARR47 pKa = 11.84RR48 pKa = 11.84ARR50 pKa = 11.84RR51 pKa = 11.84GAGAGVARR59 pKa = 11.84ARR61 pKa = 11.84GGARR65 pKa = 11.84GGQQRR70 pKa = 11.84RR71 pKa = 11.84AGRR74 pKa = 11.84GRR76 pKa = 11.84LL77 pKa = 3.45
Molecular weight: 8.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5818762 |
47 |
7172 |
347.6 |
36.43 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
16.585 ± 0.051 | 1.491 ± 0.01 |
4.715 ± 0.014 | 5.353 ± 0.027 |
2.442 ± 0.013 | 10.513 ± 0.031 |
1.94 ± 0.01 | 2.347 ± 0.012 |
2.865 ± 0.018 | 9.499 ± 0.027 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.785 ± 0.009 | 1.88 ± 0.014 |
6.405 ± 0.023 | 5.607 ± 0.038 |
6.928 ± 0.022 | 6.338 ± 0.018 |
4.059 ± 0.019 | 6.247 ± 0.018 |
1.341 ± 0.008 | 1.663 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
