
Caulobacteraceae bacterium OTSz_A_272
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Caulobacterales; Caulobacteraceae; unclassified Caulobacteraceae
Average proteome isoelectric point is 6.42
Get precalculated fractions of proteins
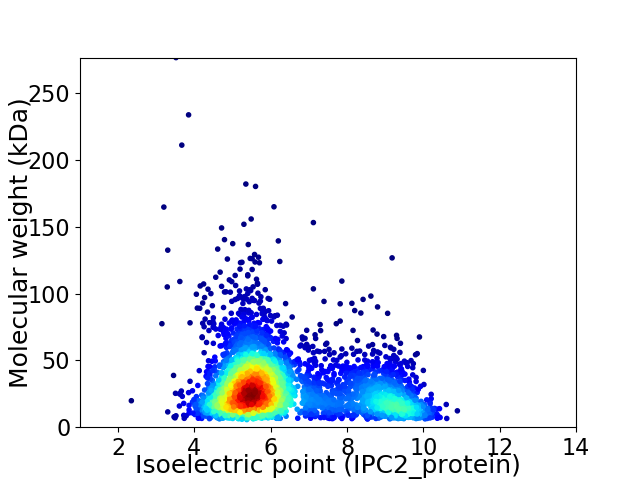
Virtual 2D-PAGE plot for 3743 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1B1AKB8|A0A1B1AKB8_9CAUL FolB domain-containing protein OS=Caulobacteraceae bacterium OTSz_A_272 OX=1759059 GN=ATE48_14310 PE=4 SV=1
MM1 pKa = 7.65RR2 pKa = 11.84NWIIGAAALLAVAAPSVAAAQTGYY26 pKa = 11.2VGVNYY31 pKa = 10.6GNVDD35 pKa = 2.94AGGGVDD41 pKa = 3.46EE42 pKa = 5.75DD43 pKa = 5.39FYY45 pKa = 11.89GLDD48 pKa = 3.44GAVAFAGSGSIVFEE62 pKa = 3.67VDD64 pKa = 2.88AAVVDD69 pKa = 4.79SDD71 pKa = 4.98DD72 pKa = 3.43TDD74 pKa = 3.04TGYY77 pKa = 11.79GLTGHH82 pKa = 6.59VYY84 pKa = 10.52SRR86 pKa = 11.84NDD88 pKa = 2.74SHH90 pKa = 8.46LFGGFIGVADD100 pKa = 4.49SNDD103 pKa = 3.08STTWVGGLEE112 pKa = 3.87AAKK115 pKa = 10.72YY116 pKa = 5.31YY117 pKa = 9.75TNWTLAGAVFYY128 pKa = 11.24GNNDD132 pKa = 3.59DD133 pKa = 5.55ADD135 pKa = 3.64VDD137 pKa = 4.57GYY139 pKa = 10.98GLNVEE144 pKa = 4.35ARR146 pKa = 11.84AFVSDD151 pKa = 3.88NFRR154 pKa = 11.84LNANIGYY161 pKa = 10.47ANVDD165 pKa = 3.28AGAGDD170 pKa = 4.87DD171 pKa = 3.8DD172 pKa = 5.26AMIYY176 pKa = 10.54GVGGEE181 pKa = 4.18YY182 pKa = 10.34QFAAMPISIVAGYY195 pKa = 7.92STTDD199 pKa = 3.55FDD201 pKa = 5.37GGDD204 pKa = 3.61VDD206 pKa = 3.97TLSIGVRR213 pKa = 11.84YY214 pKa = 9.61NWGGTLRR221 pKa = 11.84DD222 pKa = 3.72RR223 pKa = 11.84DD224 pKa = 3.47RR225 pKa = 11.84SGASQASLTGVGSVFF240 pKa = 3.69
MM1 pKa = 7.65RR2 pKa = 11.84NWIIGAAALLAVAAPSVAAAQTGYY26 pKa = 11.2VGVNYY31 pKa = 10.6GNVDD35 pKa = 2.94AGGGVDD41 pKa = 3.46EE42 pKa = 5.75DD43 pKa = 5.39FYY45 pKa = 11.89GLDD48 pKa = 3.44GAVAFAGSGSIVFEE62 pKa = 3.67VDD64 pKa = 2.88AAVVDD69 pKa = 4.79SDD71 pKa = 4.98DD72 pKa = 3.43TDD74 pKa = 3.04TGYY77 pKa = 11.79GLTGHH82 pKa = 6.59VYY84 pKa = 10.52SRR86 pKa = 11.84NDD88 pKa = 2.74SHH90 pKa = 8.46LFGGFIGVADD100 pKa = 4.49SNDD103 pKa = 3.08STTWVGGLEE112 pKa = 3.87AAKK115 pKa = 10.72YY116 pKa = 5.31YY117 pKa = 9.75TNWTLAGAVFYY128 pKa = 11.24GNNDD132 pKa = 3.59DD133 pKa = 5.55ADD135 pKa = 3.64VDD137 pKa = 4.57GYY139 pKa = 10.98GLNVEE144 pKa = 4.35ARR146 pKa = 11.84AFVSDD151 pKa = 3.88NFRR154 pKa = 11.84LNANIGYY161 pKa = 10.47ANVDD165 pKa = 3.28AGAGDD170 pKa = 4.87DD171 pKa = 3.8DD172 pKa = 5.26AMIYY176 pKa = 10.54GVGGEE181 pKa = 4.18YY182 pKa = 10.34QFAAMPISIVAGYY195 pKa = 7.92STTDD199 pKa = 3.55FDD201 pKa = 5.37GGDD204 pKa = 3.61VDD206 pKa = 3.97TLSIGVRR213 pKa = 11.84YY214 pKa = 9.61NWGGTLRR221 pKa = 11.84DD222 pKa = 3.72RR223 pKa = 11.84DD224 pKa = 3.47RR225 pKa = 11.84SGASQASLTGVGSVFF240 pKa = 3.69
Molecular weight: 24.72 kDa
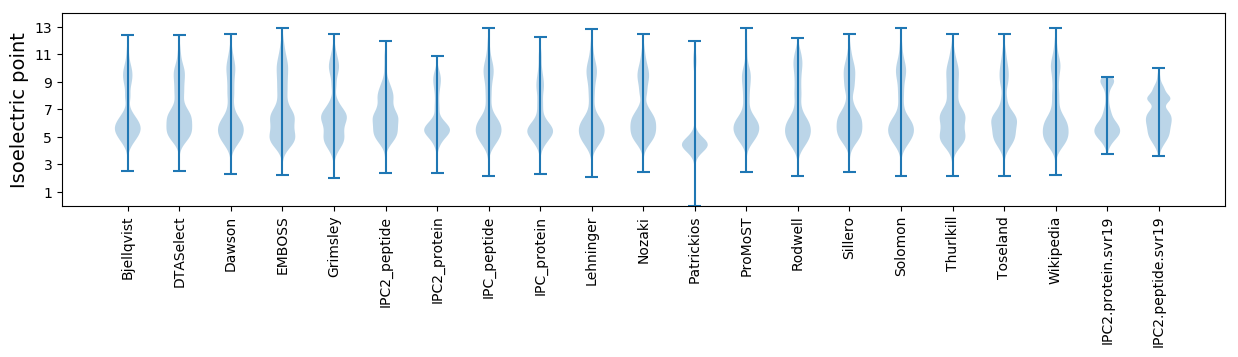
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1B1AJZ8|A0A1B1AJZ8_9CAUL MFS transporter OS=Caulobacteraceae bacterium OTSz_A_272 OX=1759059 GN=ATE48_13710 PE=4 SV=1
MM1 pKa = 7.68KK2 pKa = 10.61LNLLHH7 pKa = 6.94RR8 pKa = 11.84WFGALVEE15 pKa = 4.54SDD17 pKa = 3.59DD18 pKa = 4.99HH19 pKa = 7.03GVTLLRR25 pKa = 11.84FLAVGGTATLVMMLLSISFSSGFGFPPQVAQGVAHH60 pKa = 7.41ALCIVPTYY68 pKa = 10.27LCQRR72 pKa = 11.84AITFRR77 pKa = 11.84SNVQHH82 pKa = 6.88RR83 pKa = 11.84RR84 pKa = 11.84GLLGYY89 pKa = 10.8LSMQLPLLGMGVALAWLLIGQMHH112 pKa = 7.08WPRR115 pKa = 11.84EE116 pKa = 3.88IGLATIAVIVGATSFLVQRR135 pKa = 11.84LVIFASKK142 pKa = 9.51SQKK145 pKa = 10.52
MM1 pKa = 7.68KK2 pKa = 10.61LNLLHH7 pKa = 6.94RR8 pKa = 11.84WFGALVEE15 pKa = 4.54SDD17 pKa = 3.59DD18 pKa = 4.99HH19 pKa = 7.03GVTLLRR25 pKa = 11.84FLAVGGTATLVMMLLSISFSSGFGFPPQVAQGVAHH60 pKa = 7.41ALCIVPTYY68 pKa = 10.27LCQRR72 pKa = 11.84AITFRR77 pKa = 11.84SNVQHH82 pKa = 6.88RR83 pKa = 11.84RR84 pKa = 11.84GLLGYY89 pKa = 10.8LSMQLPLLGMGVALAWLLIGQMHH112 pKa = 7.08WPRR115 pKa = 11.84EE116 pKa = 3.88IGLATIAVIVGATSFLVQRR135 pKa = 11.84LVIFASKK142 pKa = 9.51SQKK145 pKa = 10.52
Molecular weight: 15.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1113520 |
53 |
2768 |
297.5 |
32.24 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.662 ± 0.065 | 0.804 ± 0.014 |
5.661 ± 0.034 | 5.892 ± 0.032 |
3.798 ± 0.023 | 8.47 ± 0.051 |
1.895 ± 0.019 | 5.029 ± 0.029 |
2.911 ± 0.048 | 9.582 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.347 ± 0.02 | 2.821 ± 0.027 |
5.074 ± 0.034 | 3.367 ± 0.026 |
7.461 ± 0.044 | 5.358 ± 0.029 |
5.112 ± 0.039 | 7.121 ± 0.031 |
1.433 ± 0.017 | 2.204 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
