
Xinfangfangia humi
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Xinfangfangia
Average proteome isoelectric point is 6.5
Get precalculated fractions of proteins
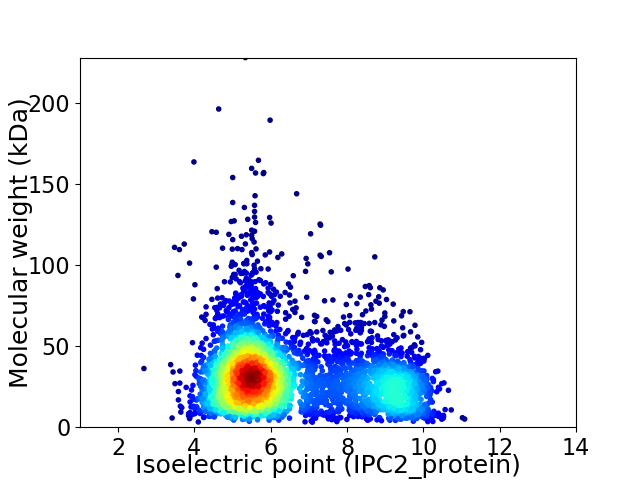
Virtual 2D-PAGE plot for 4199 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3P5XEK8|A0A3P5XEK8_9RHOB Inner membrane protein YdcO OS=Xinfangfangia humi OX=2483812 GN=ydcO PE=4 SV=1
MM1 pKa = 7.16ATINGTAGNDD11 pKa = 3.59TLISGDD17 pKa = 3.96QADD20 pKa = 4.3TLNGLGGDD28 pKa = 3.75DD29 pKa = 3.58VLYY32 pKa = 11.09GGGGNDD38 pKa = 3.68SLSGGDD44 pKa = 4.98GNDD47 pKa = 3.54LLYY50 pKa = 11.22GGDD53 pKa = 4.8GNDD56 pKa = 3.8TLNGGNGNDD65 pKa = 3.94TLIGGPGADD74 pKa = 3.88SLIGGAGIDD83 pKa = 3.57TVDD86 pKa = 3.54YY87 pKa = 9.8SGSGAAVNVNLGSSSAATGGDD108 pKa = 3.46AQGDD112 pKa = 3.97RR113 pKa = 11.84YY114 pKa = 10.98SGIEE118 pKa = 3.8IVIGSAFNDD127 pKa = 4.54TITGDD132 pKa = 3.31TGANTLSGGLGNDD145 pKa = 3.25QLYY148 pKa = 10.69GGAGADD154 pKa = 3.5VLDD157 pKa = 4.55GGEE160 pKa = 4.8GNDD163 pKa = 3.59SLYY166 pKa = 11.17GGSGNDD172 pKa = 3.62TLIGGAGADD181 pKa = 3.97YY182 pKa = 11.08LDD184 pKa = 4.86GGADD188 pKa = 3.82SDD190 pKa = 4.24TVDD193 pKa = 3.74YY194 pKa = 10.8SGSDD198 pKa = 2.93AAVNVNLTSGSGSGGHH214 pKa = 6.16AQGDD218 pKa = 4.05TYY220 pKa = 11.88VNIEE224 pKa = 4.56NITGSDD230 pKa = 3.72YY231 pKa = 11.76DD232 pKa = 4.49DD233 pKa = 3.84VLIGNSGANILSGGAGNDD251 pKa = 3.27SLYY254 pKa = 11.29GGGGNDD260 pKa = 3.28SLYY263 pKa = 11.28GGDD266 pKa = 5.58GDD268 pKa = 5.68DD269 pKa = 5.6LLDD272 pKa = 4.68GGDD275 pKa = 4.63GNDD278 pKa = 3.56SLYY281 pKa = 11.22GGNGNDD287 pKa = 3.82TLIGGAGSDD296 pKa = 3.98RR297 pKa = 11.84LDD299 pKa = 3.58GGAGIDD305 pKa = 3.55TVDD308 pKa = 3.59YY309 pKa = 10.77SGSTSGVNVNLSTGSGSSGHH329 pKa = 6.78AAGDD333 pKa = 3.74TYY335 pKa = 11.69VSIEE339 pKa = 4.0NVIGSAYY346 pKa = 10.51NDD348 pKa = 4.31TIVGTAGANYY358 pKa = 10.13LSGGAGRR365 pKa = 11.84DD366 pKa = 3.83SISGGDD372 pKa = 4.08GDD374 pKa = 4.44DD375 pKa = 3.81TLAGGAGADD384 pKa = 4.06TLNGGQGLDD393 pKa = 2.88IVDD396 pKa = 3.96YY397 pKa = 10.58SEE399 pKa = 5.04SDD401 pKa = 3.09AAVNINLSTWTASGGHH417 pKa = 5.47ATGDD421 pKa = 3.68VLSGIDD427 pKa = 4.38GIIGSAFDD435 pKa = 4.04DD436 pKa = 3.75VLIGFNEE443 pKa = 4.25MGTGADD449 pKa = 3.18SYY451 pKa = 11.4TNVIYY456 pKa = 10.59GGAGNDD462 pKa = 4.13TIDD465 pKa = 4.55GMGGDD470 pKa = 4.1DD471 pKa = 3.96SLYY474 pKa = 10.9GGTGNDD480 pKa = 3.5YY481 pKa = 11.52VLGGSGNDD489 pKa = 3.16TVYY492 pKa = 11.2GDD494 pKa = 3.84EE495 pKa = 4.59GDD497 pKa = 3.7DD498 pKa = 3.23RR499 pKa = 11.84VYY501 pKa = 11.13GGSGDD506 pKa = 4.2DD507 pKa = 3.88LVSGGSGNDD516 pKa = 3.06QVYY519 pKa = 10.77GDD521 pKa = 4.77EE522 pKa = 4.68GTDD525 pKa = 3.19TLYY528 pKa = 11.25GDD530 pKa = 4.06EE531 pKa = 5.04GNDD534 pKa = 3.28VLYY537 pKa = 11.02GGTGNDD543 pKa = 3.93LLYY546 pKa = 11.19GGADD550 pKa = 3.57EE551 pKa = 4.53DD552 pKa = 4.18TLYY555 pKa = 11.36GGDD558 pKa = 4.52GVDD561 pKa = 3.79TLHH564 pKa = 7.13GDD566 pKa = 3.44SGNDD570 pKa = 3.43SLFGDD575 pKa = 4.51AGNDD579 pKa = 3.5LLYY582 pKa = 11.04GGTGDD587 pKa = 3.51DD588 pKa = 3.76TLYY591 pKa = 11.28GGAGNDD597 pKa = 3.4QLFGGDD603 pKa = 3.57GRR605 pKa = 11.84DD606 pKa = 3.48VLHH609 pKa = 7.49ADD611 pKa = 4.14GGNDD615 pKa = 3.42TLTGGAGNDD624 pKa = 3.46VFVVYY629 pKa = 9.9PGSGAVLITDD639 pKa = 4.28FGAGNTGPIDD649 pKa = 5.67DD650 pKa = 5.87DD651 pKa = 5.61DD652 pKa = 3.92NTNNDD657 pKa = 3.86FVDD660 pKa = 4.73LSAWYY665 pKa = 10.74NEE667 pKa = 4.0TTLAAWNAANPGQTYY682 pKa = 10.74NNPLQWMIADD692 pKa = 3.98HH693 pKa = 7.41ADD695 pKa = 3.29GVLNAVGGGLRR706 pKa = 11.84IQNGGVAVSSDD717 pKa = 3.18MLVFEE722 pKa = 4.65TTGVVCFARR731 pKa = 11.84GTRR734 pKa = 11.84ILTPRR739 pKa = 11.84GEE741 pKa = 4.28IAVEE745 pKa = 3.61ALRR748 pKa = 11.84AGDD751 pKa = 4.9LVTTLDD757 pKa = 4.2AGPQVLRR764 pKa = 11.84WIGSRR769 pKa = 11.84RR770 pKa = 11.84IGAAEE775 pKa = 4.05LAADD779 pKa = 4.33PGLIPVRR786 pKa = 11.84IRR788 pKa = 11.84AGSLGQGLPRR798 pKa = 11.84RR799 pKa = 11.84DD800 pKa = 3.37LTVSPQHH807 pKa = 6.83RR808 pKa = 11.84ILLRR812 pKa = 11.84SRR814 pKa = 11.84IVEE817 pKa = 4.04RR818 pKa = 11.84MTGEE822 pKa = 4.2PEE824 pKa = 3.72VLLAAKK830 pKa = 10.32HH831 pKa = 4.28MTGLDD836 pKa = 3.38GLEE839 pKa = 4.1VVGDD843 pKa = 4.01CRR845 pKa = 11.84GVEE848 pKa = 4.13YY849 pKa = 10.36FHH851 pKa = 8.04LLFDD855 pKa = 3.75RR856 pKa = 11.84HH857 pKa = 6.11QILFAEE863 pKa = 5.3GAPAEE868 pKa = 4.13SLLFGKK874 pKa = 8.27EE875 pKa = 3.57ARR877 pKa = 11.84KK878 pKa = 8.48MLRR881 pKa = 11.84PAAVEE886 pKa = 4.29EE887 pKa = 4.26ILTVLPALAVAPPPPARR904 pKa = 11.84ALMDD908 pKa = 3.43GRR910 pKa = 11.84TGRR913 pKa = 11.84SLVRR917 pKa = 11.84RR918 pKa = 11.84MRR920 pKa = 11.84QNGVEE925 pKa = 4.35AICAAEE931 pKa = 4.14APPVRR936 pKa = 11.84VTAA939 pKa = 4.38
MM1 pKa = 7.16ATINGTAGNDD11 pKa = 3.59TLISGDD17 pKa = 3.96QADD20 pKa = 4.3TLNGLGGDD28 pKa = 3.75DD29 pKa = 3.58VLYY32 pKa = 11.09GGGGNDD38 pKa = 3.68SLSGGDD44 pKa = 4.98GNDD47 pKa = 3.54LLYY50 pKa = 11.22GGDD53 pKa = 4.8GNDD56 pKa = 3.8TLNGGNGNDD65 pKa = 3.94TLIGGPGADD74 pKa = 3.88SLIGGAGIDD83 pKa = 3.57TVDD86 pKa = 3.54YY87 pKa = 9.8SGSGAAVNVNLGSSSAATGGDD108 pKa = 3.46AQGDD112 pKa = 3.97RR113 pKa = 11.84YY114 pKa = 10.98SGIEE118 pKa = 3.8IVIGSAFNDD127 pKa = 4.54TITGDD132 pKa = 3.31TGANTLSGGLGNDD145 pKa = 3.25QLYY148 pKa = 10.69GGAGADD154 pKa = 3.5VLDD157 pKa = 4.55GGEE160 pKa = 4.8GNDD163 pKa = 3.59SLYY166 pKa = 11.17GGSGNDD172 pKa = 3.62TLIGGAGADD181 pKa = 3.97YY182 pKa = 11.08LDD184 pKa = 4.86GGADD188 pKa = 3.82SDD190 pKa = 4.24TVDD193 pKa = 3.74YY194 pKa = 10.8SGSDD198 pKa = 2.93AAVNVNLTSGSGSGGHH214 pKa = 6.16AQGDD218 pKa = 4.05TYY220 pKa = 11.88VNIEE224 pKa = 4.56NITGSDD230 pKa = 3.72YY231 pKa = 11.76DD232 pKa = 4.49DD233 pKa = 3.84VLIGNSGANILSGGAGNDD251 pKa = 3.27SLYY254 pKa = 11.29GGGGNDD260 pKa = 3.28SLYY263 pKa = 11.28GGDD266 pKa = 5.58GDD268 pKa = 5.68DD269 pKa = 5.6LLDD272 pKa = 4.68GGDD275 pKa = 4.63GNDD278 pKa = 3.56SLYY281 pKa = 11.22GGNGNDD287 pKa = 3.82TLIGGAGSDD296 pKa = 3.98RR297 pKa = 11.84LDD299 pKa = 3.58GGAGIDD305 pKa = 3.55TVDD308 pKa = 3.59YY309 pKa = 10.77SGSTSGVNVNLSTGSGSSGHH329 pKa = 6.78AAGDD333 pKa = 3.74TYY335 pKa = 11.69VSIEE339 pKa = 4.0NVIGSAYY346 pKa = 10.51NDD348 pKa = 4.31TIVGTAGANYY358 pKa = 10.13LSGGAGRR365 pKa = 11.84DD366 pKa = 3.83SISGGDD372 pKa = 4.08GDD374 pKa = 4.44DD375 pKa = 3.81TLAGGAGADD384 pKa = 4.06TLNGGQGLDD393 pKa = 2.88IVDD396 pKa = 3.96YY397 pKa = 10.58SEE399 pKa = 5.04SDD401 pKa = 3.09AAVNINLSTWTASGGHH417 pKa = 5.47ATGDD421 pKa = 3.68VLSGIDD427 pKa = 4.38GIIGSAFDD435 pKa = 4.04DD436 pKa = 3.75VLIGFNEE443 pKa = 4.25MGTGADD449 pKa = 3.18SYY451 pKa = 11.4TNVIYY456 pKa = 10.59GGAGNDD462 pKa = 4.13TIDD465 pKa = 4.55GMGGDD470 pKa = 4.1DD471 pKa = 3.96SLYY474 pKa = 10.9GGTGNDD480 pKa = 3.5YY481 pKa = 11.52VLGGSGNDD489 pKa = 3.16TVYY492 pKa = 11.2GDD494 pKa = 3.84EE495 pKa = 4.59GDD497 pKa = 3.7DD498 pKa = 3.23RR499 pKa = 11.84VYY501 pKa = 11.13GGSGDD506 pKa = 4.2DD507 pKa = 3.88LVSGGSGNDD516 pKa = 3.06QVYY519 pKa = 10.77GDD521 pKa = 4.77EE522 pKa = 4.68GTDD525 pKa = 3.19TLYY528 pKa = 11.25GDD530 pKa = 4.06EE531 pKa = 5.04GNDD534 pKa = 3.28VLYY537 pKa = 11.02GGTGNDD543 pKa = 3.93LLYY546 pKa = 11.19GGADD550 pKa = 3.57EE551 pKa = 4.53DD552 pKa = 4.18TLYY555 pKa = 11.36GGDD558 pKa = 4.52GVDD561 pKa = 3.79TLHH564 pKa = 7.13GDD566 pKa = 3.44SGNDD570 pKa = 3.43SLFGDD575 pKa = 4.51AGNDD579 pKa = 3.5LLYY582 pKa = 11.04GGTGDD587 pKa = 3.51DD588 pKa = 3.76TLYY591 pKa = 11.28GGAGNDD597 pKa = 3.4QLFGGDD603 pKa = 3.57GRR605 pKa = 11.84DD606 pKa = 3.48VLHH609 pKa = 7.49ADD611 pKa = 4.14GGNDD615 pKa = 3.42TLTGGAGNDD624 pKa = 3.46VFVVYY629 pKa = 9.9PGSGAVLITDD639 pKa = 4.28FGAGNTGPIDD649 pKa = 5.67DD650 pKa = 5.87DD651 pKa = 5.61DD652 pKa = 3.92NTNNDD657 pKa = 3.86FVDD660 pKa = 4.73LSAWYY665 pKa = 10.74NEE667 pKa = 4.0TTLAAWNAANPGQTYY682 pKa = 10.74NNPLQWMIADD692 pKa = 3.98HH693 pKa = 7.41ADD695 pKa = 3.29GVLNAVGGGLRR706 pKa = 11.84IQNGGVAVSSDD717 pKa = 3.18MLVFEE722 pKa = 4.65TTGVVCFARR731 pKa = 11.84GTRR734 pKa = 11.84ILTPRR739 pKa = 11.84GEE741 pKa = 4.28IAVEE745 pKa = 3.61ALRR748 pKa = 11.84AGDD751 pKa = 4.9LVTTLDD757 pKa = 4.2AGPQVLRR764 pKa = 11.84WIGSRR769 pKa = 11.84RR770 pKa = 11.84IGAAEE775 pKa = 4.05LAADD779 pKa = 4.33PGLIPVRR786 pKa = 11.84IRR788 pKa = 11.84AGSLGQGLPRR798 pKa = 11.84RR799 pKa = 11.84DD800 pKa = 3.37LTVSPQHH807 pKa = 6.83RR808 pKa = 11.84ILLRR812 pKa = 11.84SRR814 pKa = 11.84IVEE817 pKa = 4.04RR818 pKa = 11.84MTGEE822 pKa = 4.2PEE824 pKa = 3.72VLLAAKK830 pKa = 10.32HH831 pKa = 4.28MTGLDD836 pKa = 3.38GLEE839 pKa = 4.1VVGDD843 pKa = 4.01CRR845 pKa = 11.84GVEE848 pKa = 4.13YY849 pKa = 10.36FHH851 pKa = 8.04LLFDD855 pKa = 3.75RR856 pKa = 11.84HH857 pKa = 6.11QILFAEE863 pKa = 5.3GAPAEE868 pKa = 4.13SLLFGKK874 pKa = 8.27EE875 pKa = 3.57ARR877 pKa = 11.84KK878 pKa = 8.48MLRR881 pKa = 11.84PAAVEE886 pKa = 4.29EE887 pKa = 4.26ILTVLPALAVAPPPPARR904 pKa = 11.84ALMDD908 pKa = 3.43GRR910 pKa = 11.84TGRR913 pKa = 11.84SLVRR917 pKa = 11.84RR918 pKa = 11.84MRR920 pKa = 11.84QNGVEE925 pKa = 4.35AICAAEE931 pKa = 4.14APPVRR936 pKa = 11.84VTAA939 pKa = 4.38
Molecular weight: 93.53 kDa
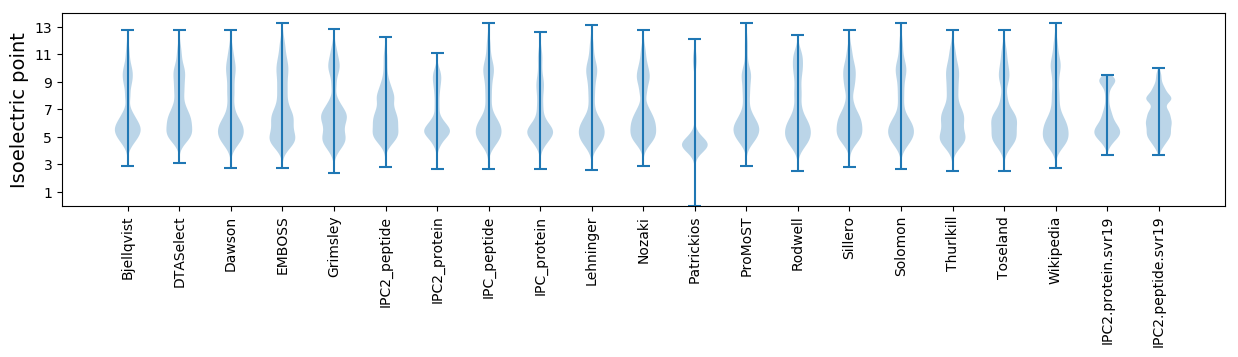
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3P5WSR9|A0A3P5WSR9_9RHOB Sarcosine oxidase gamma subunit family OS=Xinfangfangia humi OX=2483812 GN=XINFAN_01672 PE=4 SV=1
MM1 pKa = 7.36RR2 pKa = 11.84RR3 pKa = 11.84VRR5 pKa = 11.84MGAAPRR11 pKa = 11.84APRR14 pKa = 11.84PLWKK18 pKa = 10.22RR19 pKa = 11.84LGWFALIWALSVAALGLVALVFRR42 pKa = 11.84LMMKK46 pKa = 10.6LAGLSLL52 pKa = 3.8
MM1 pKa = 7.36RR2 pKa = 11.84RR3 pKa = 11.84VRR5 pKa = 11.84MGAAPRR11 pKa = 11.84APRR14 pKa = 11.84PLWKK18 pKa = 10.22RR19 pKa = 11.84LGWFALIWALSVAALGLVALVFRR42 pKa = 11.84LMMKK46 pKa = 10.6LAGLSLL52 pKa = 3.8
Molecular weight: 5.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1295700 |
29 |
2121 |
308.6 |
33.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.275 ± 0.058 | 0.881 ± 0.01 |
5.526 ± 0.031 | 5.918 ± 0.032 |
3.566 ± 0.023 | 9.217 ± 0.038 |
1.996 ± 0.018 | 4.957 ± 0.025 |
2.683 ± 0.031 | 10.502 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.599 ± 0.018 | 2.261 ± 0.021 |
5.548 ± 0.032 | 2.949 ± 0.019 |
7.605 ± 0.043 | 4.97 ± 0.024 |
5.03 ± 0.026 | 6.976 ± 0.031 |
1.504 ± 0.016 | 2.035 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
