
Salipaludibacillus neizhouensis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillaceae; Salipaludibacillus
Average proteome isoelectric point is 6.14
Get precalculated fractions of proteins
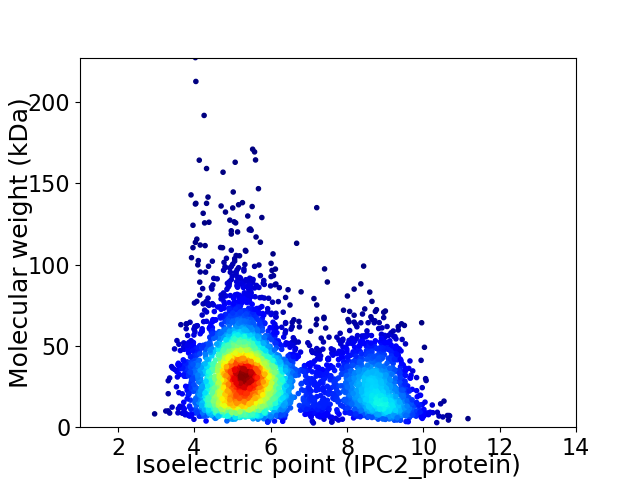
Virtual 2D-PAGE plot for 4767 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3A9KBN9|A0A3A9KBN9_9BACI LMWPc domain-containing protein OS=Salipaludibacillus neizhouensis OX=885475 GN=CR203_24580 PE=4 SV=1
MM1 pKa = 7.5KK2 pKa = 10.29KK3 pKa = 9.41ILLIIGSSLLVALVSACGSNNEE25 pKa = 4.16VSNAGNNTNQSSSDD39 pKa = 3.31QTLTVLVEE47 pKa = 4.52GGSPAQTVADD57 pKa = 3.77LTKK60 pKa = 11.06EE61 pKa = 3.94EE62 pKa = 4.49FEE64 pKa = 4.59EE65 pKa = 4.11NTGYY69 pKa = 10.72EE70 pKa = 4.36VIIEE74 pKa = 4.26SVPYY78 pKa = 10.06SGVYY82 pKa = 10.55DD83 pKa = 3.98RR84 pKa = 11.84MRR86 pKa = 11.84TEE88 pKa = 4.07LTSGAGAFDD97 pKa = 3.72VATIDD102 pKa = 4.5TIWLPALYY110 pKa = 9.93QGLEE114 pKa = 4.31PIQEE118 pKa = 4.45EE119 pKa = 4.51ITDD122 pKa = 3.83EE123 pKa = 4.17MQDD126 pKa = 3.61DD127 pKa = 5.18LFPGLIDD134 pKa = 3.97GASMDD139 pKa = 3.76GNVYY143 pKa = 10.61GLPTWTNSKK152 pKa = 9.89ILLYY156 pKa = 10.65RR157 pKa = 11.84EE158 pKa = 4.69DD159 pKa = 5.07LFNDD163 pKa = 3.76SNNQDD168 pKa = 3.05EE169 pKa = 4.71FLDD172 pKa = 3.69EE173 pKa = 4.71FGYY176 pKa = 10.64EE177 pKa = 4.09LSPPEE182 pKa = 3.87TWDD185 pKa = 3.09QYY187 pKa = 11.82KK188 pKa = 10.69DD189 pKa = 3.38VAHH192 pKa = 6.69FFGNNEE198 pKa = 4.44NYY200 pKa = 9.77PEE202 pKa = 4.52LYY204 pKa = 8.15GTSVFGATSGDD215 pKa = 4.25SVASWLDD222 pKa = 3.3HH223 pKa = 5.53VAQAGAEE230 pKa = 3.81ALVINDD236 pKa = 3.56AGEE239 pKa = 4.32VIVNDD244 pKa = 4.41DD245 pKa = 3.62DD246 pKa = 4.66HH247 pKa = 7.67VAALEE252 pKa = 3.9FLNDD256 pKa = 3.67LVNEE260 pKa = 4.22EE261 pKa = 4.4GVVPEE266 pKa = 4.77GVLEE270 pKa = 4.13TASAEE275 pKa = 4.21TSEE278 pKa = 5.0LFYY281 pKa = 11.31NGNLAMMLAWGHH293 pKa = 5.91FFVPSNDD300 pKa = 3.53PDD302 pKa = 3.8TSDD305 pKa = 3.17VAGDD309 pKa = 3.37VGAAPMIAGDD319 pKa = 3.8GGIGVVPGPWYY330 pKa = 10.32QVIPSSSEE338 pKa = 3.63KK339 pKa = 10.38KK340 pKa = 10.63DD341 pKa = 3.12IAKK344 pKa = 10.24EE345 pKa = 3.84YY346 pKa = 11.26LLFLYY351 pKa = 9.96EE352 pKa = 4.13NNEE355 pKa = 4.13LYY357 pKa = 11.04ADD359 pKa = 4.08ALGVAARR366 pKa = 11.84QSVFEE371 pKa = 4.53EE372 pKa = 4.23YY373 pKa = 10.71GQQEE377 pKa = 4.84EE378 pKa = 4.69YY379 pKa = 11.56AHH381 pKa = 6.6LNEE384 pKa = 5.65LATTLNGKK392 pKa = 7.08QTQNRR397 pKa = 11.84PSTEE401 pKa = 3.22KK402 pKa = 9.79WEE404 pKa = 4.56EE405 pKa = 4.0IEE407 pKa = 5.65SEE409 pKa = 4.19ALLPAVQNVFSGSQTPQEE427 pKa = 4.51ALDD430 pKa = 3.75EE431 pKa = 4.43AKK433 pKa = 10.51EE434 pKa = 4.01IIEE437 pKa = 4.83GIVNN441 pKa = 3.98
MM1 pKa = 7.5KK2 pKa = 10.29KK3 pKa = 9.41ILLIIGSSLLVALVSACGSNNEE25 pKa = 4.16VSNAGNNTNQSSSDD39 pKa = 3.31QTLTVLVEE47 pKa = 4.52GGSPAQTVADD57 pKa = 3.77LTKK60 pKa = 11.06EE61 pKa = 3.94EE62 pKa = 4.49FEE64 pKa = 4.59EE65 pKa = 4.11NTGYY69 pKa = 10.72EE70 pKa = 4.36VIIEE74 pKa = 4.26SVPYY78 pKa = 10.06SGVYY82 pKa = 10.55DD83 pKa = 3.98RR84 pKa = 11.84MRR86 pKa = 11.84TEE88 pKa = 4.07LTSGAGAFDD97 pKa = 3.72VATIDD102 pKa = 4.5TIWLPALYY110 pKa = 9.93QGLEE114 pKa = 4.31PIQEE118 pKa = 4.45EE119 pKa = 4.51ITDD122 pKa = 3.83EE123 pKa = 4.17MQDD126 pKa = 3.61DD127 pKa = 5.18LFPGLIDD134 pKa = 3.97GASMDD139 pKa = 3.76GNVYY143 pKa = 10.61GLPTWTNSKK152 pKa = 9.89ILLYY156 pKa = 10.65RR157 pKa = 11.84EE158 pKa = 4.69DD159 pKa = 5.07LFNDD163 pKa = 3.76SNNQDD168 pKa = 3.05EE169 pKa = 4.71FLDD172 pKa = 3.69EE173 pKa = 4.71FGYY176 pKa = 10.64EE177 pKa = 4.09LSPPEE182 pKa = 3.87TWDD185 pKa = 3.09QYY187 pKa = 11.82KK188 pKa = 10.69DD189 pKa = 3.38VAHH192 pKa = 6.69FFGNNEE198 pKa = 4.44NYY200 pKa = 9.77PEE202 pKa = 4.52LYY204 pKa = 8.15GTSVFGATSGDD215 pKa = 4.25SVASWLDD222 pKa = 3.3HH223 pKa = 5.53VAQAGAEE230 pKa = 3.81ALVINDD236 pKa = 3.56AGEE239 pKa = 4.32VIVNDD244 pKa = 4.41DD245 pKa = 3.62DD246 pKa = 4.66HH247 pKa = 7.67VAALEE252 pKa = 3.9FLNDD256 pKa = 3.67LVNEE260 pKa = 4.22EE261 pKa = 4.4GVVPEE266 pKa = 4.77GVLEE270 pKa = 4.13TASAEE275 pKa = 4.21TSEE278 pKa = 5.0LFYY281 pKa = 11.31NGNLAMMLAWGHH293 pKa = 5.91FFVPSNDD300 pKa = 3.53PDD302 pKa = 3.8TSDD305 pKa = 3.17VAGDD309 pKa = 3.37VGAAPMIAGDD319 pKa = 3.8GGIGVVPGPWYY330 pKa = 10.32QVIPSSSEE338 pKa = 3.63KK339 pKa = 10.38KK340 pKa = 10.63DD341 pKa = 3.12IAKK344 pKa = 10.24EE345 pKa = 3.84YY346 pKa = 11.26LLFLYY351 pKa = 9.96EE352 pKa = 4.13NNEE355 pKa = 4.13LYY357 pKa = 11.04ADD359 pKa = 4.08ALGVAARR366 pKa = 11.84QSVFEE371 pKa = 4.53EE372 pKa = 4.23YY373 pKa = 10.71GQQEE377 pKa = 4.84EE378 pKa = 4.69YY379 pKa = 11.56AHH381 pKa = 6.6LNEE384 pKa = 5.65LATTLNGKK392 pKa = 7.08QTQNRR397 pKa = 11.84PSTEE401 pKa = 3.22KK402 pKa = 9.79WEE404 pKa = 4.56EE405 pKa = 4.0IEE407 pKa = 5.65SEE409 pKa = 4.19ALLPAVQNVFSGSQTPQEE427 pKa = 4.51ALDD430 pKa = 3.75EE431 pKa = 4.43AKK433 pKa = 10.51EE434 pKa = 4.01IIEE437 pKa = 4.83GIVNN441 pKa = 3.98
Molecular weight: 47.93 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3A9JYW1|A0A3A9JYW1_9BACI Iron-siderophore ABC transporter permease OS=Salipaludibacillus neizhouensis OX=885475 GN=CR203_20735 PE=3 SV=1
MM1 pKa = 7.69GKK3 pKa = 7.98PTFNPNNRR11 pKa = 11.84KK12 pKa = 9.34RR13 pKa = 11.84KK14 pKa = 8.34KK15 pKa = 9.1NHH17 pKa = 4.73GFRR20 pKa = 11.84ARR22 pKa = 11.84MATKK26 pKa = 10.24NGRR29 pKa = 11.84HH30 pKa = 5.46ILANRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 8.72GRR40 pKa = 11.84KK41 pKa = 8.69VLSAA45 pKa = 4.05
MM1 pKa = 7.69GKK3 pKa = 7.98PTFNPNNRR11 pKa = 11.84KK12 pKa = 9.34RR13 pKa = 11.84KK14 pKa = 8.34KK15 pKa = 9.1NHH17 pKa = 4.73GFRR20 pKa = 11.84ARR22 pKa = 11.84MATKK26 pKa = 10.24NGRR29 pKa = 11.84HH30 pKa = 5.46ILANRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 8.72GRR40 pKa = 11.84KK41 pKa = 8.69VLSAA45 pKa = 4.05
Molecular weight: 5.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1414254 |
23 |
2034 |
296.7 |
33.39 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.402 ± 0.036 | 0.668 ± 0.011 |
5.376 ± 0.03 | 7.856 ± 0.044 |
4.664 ± 0.028 | 6.805 ± 0.034 |
2.121 ± 0.017 | 7.914 ± 0.039 |
6.62 ± 0.036 | 9.656 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.822 ± 0.019 | 4.672 ± 0.027 |
3.498 ± 0.02 | 3.562 ± 0.021 |
3.919 ± 0.024 | 6.442 ± 0.026 |
5.483 ± 0.021 | 6.914 ± 0.027 |
1.099 ± 0.013 | 3.505 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
