
Modestobacter sp. DSM 44400
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Geodermatophilales; Geodermatophilaceae; Modestobacter; unclassified Modestobacter
Average proteome isoelectric point is 6.32
Get precalculated fractions of proteins
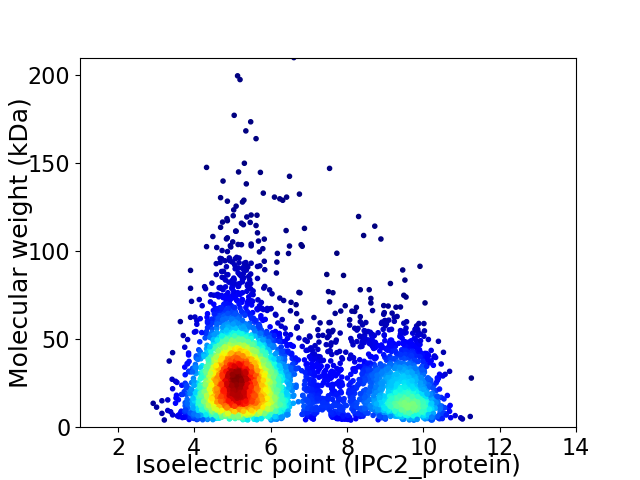
Virtual 2D-PAGE plot for 5140 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H3DL19|A0A1H3DL19_9ACTN Uncharacterized protein OS=Modestobacter sp. DSM 44400 OX=1550230 GN=SAMN05661080_00770 PE=4 SV=1
MM1 pKa = 7.82PSPSRR6 pKa = 11.84RR7 pKa = 11.84LLAAVVVVSIAALPVLAGCGVTFVVGSASPATGQVGTVQSQDD49 pKa = 3.31FPVIGATEE57 pKa = 4.07DD58 pKa = 4.48EE59 pKa = 4.25IDD61 pKa = 3.53QLARR65 pKa = 11.84NALVDD70 pKa = 6.0LNVFWQNQFPDD81 pKa = 3.34TFGQEE86 pKa = 4.06FTPLRR91 pKa = 11.84GGYY94 pKa = 10.25YY95 pKa = 10.35SVDD98 pKa = 3.64PANIDD103 pKa = 3.2PTQYY107 pKa = 10.3PGGQIGCGEE116 pKa = 4.2TPDD119 pKa = 4.31AVEE122 pKa = 5.72DD123 pKa = 3.69NAFYY127 pKa = 10.75CPPSNEE133 pKa = 4.22YY134 pKa = 10.38PNGDD138 pKa = 3.94AIQYY142 pKa = 10.28DD143 pKa = 3.93RR144 pKa = 11.84AFLQDD149 pKa = 3.64LANEE153 pKa = 4.26YY154 pKa = 11.16GRR156 pKa = 11.84FLPALVMAHH165 pKa = 6.3EE166 pKa = 5.34FGHH169 pKa = 7.21AIQGRR174 pKa = 11.84VGYY177 pKa = 8.71PLQASIAIEE186 pKa = 4.03TQADD190 pKa = 3.98CFAGAWTAWVADD202 pKa = 3.86GNAPHH207 pKa = 5.84TTIRR211 pKa = 11.84TPEE214 pKa = 4.36LDD216 pKa = 3.62DD217 pKa = 3.56VLRR220 pKa = 11.84GYY222 pKa = 11.18LLLRR226 pKa = 11.84DD227 pKa = 4.03PVGTDD232 pKa = 3.31LNEE235 pKa = 4.38SEE237 pKa = 4.52AHH239 pKa = 6.15GSLFDD244 pKa = 3.77RR245 pKa = 11.84VSAFQQGFDD254 pKa = 4.79DD255 pKa = 5.89GPTACRR261 pKa = 11.84DD262 pKa = 3.22EE263 pKa = 4.9FGADD267 pKa = 3.14RR268 pKa = 11.84IYY270 pKa = 10.04TQRR273 pKa = 11.84EE274 pKa = 3.83FDD276 pKa = 3.84PADD279 pKa = 3.5AATQGNASYY288 pKa = 10.3PVTQQLVTEE297 pKa = 4.34SLSTFWSRR305 pKa = 11.84TFTARR310 pKa = 11.84GEE312 pKa = 4.22TFAAPDD318 pKa = 3.55VVPFSGTAPDD328 pKa = 4.45CAGQGNEE335 pKa = 3.82QADD338 pKa = 4.13LVFCANKK345 pKa = 9.2DD346 pKa = 3.52TVGYY350 pKa = 10.87DD351 pKa = 3.07EE352 pKa = 5.02TDD354 pKa = 3.1LVRR357 pKa = 11.84PVYY360 pKa = 10.55EE361 pKa = 4.04EE362 pKa = 4.33LGDD365 pKa = 3.95YY366 pKa = 10.99AVLTAVAIPYY376 pKa = 10.18SLAARR381 pKa = 11.84DD382 pKa = 3.82QLGLSTDD389 pKa = 3.71DD390 pKa = 3.14QDD392 pKa = 3.89AARR395 pKa = 11.84SAVCLTGAFTRR406 pKa = 11.84SVLSGEE412 pKa = 4.4VNGITISPGDD422 pKa = 3.81ADD424 pKa = 3.82EE425 pKa = 4.98SVQFLLGYY433 pKa = 10.29SDD435 pKa = 5.43DD436 pKa = 4.43PDD438 pKa = 3.59VLGDD442 pKa = 3.5VGLSGFQLVDD452 pKa = 3.04VFRR455 pKa = 11.84SGVVEE460 pKa = 4.36GEE462 pKa = 4.31SSCGIGG468 pKa = 3.84
MM1 pKa = 7.82PSPSRR6 pKa = 11.84RR7 pKa = 11.84LLAAVVVVSIAALPVLAGCGVTFVVGSASPATGQVGTVQSQDD49 pKa = 3.31FPVIGATEE57 pKa = 4.07DD58 pKa = 4.48EE59 pKa = 4.25IDD61 pKa = 3.53QLARR65 pKa = 11.84NALVDD70 pKa = 6.0LNVFWQNQFPDD81 pKa = 3.34TFGQEE86 pKa = 4.06FTPLRR91 pKa = 11.84GGYY94 pKa = 10.25YY95 pKa = 10.35SVDD98 pKa = 3.64PANIDD103 pKa = 3.2PTQYY107 pKa = 10.3PGGQIGCGEE116 pKa = 4.2TPDD119 pKa = 4.31AVEE122 pKa = 5.72DD123 pKa = 3.69NAFYY127 pKa = 10.75CPPSNEE133 pKa = 4.22YY134 pKa = 10.38PNGDD138 pKa = 3.94AIQYY142 pKa = 10.28DD143 pKa = 3.93RR144 pKa = 11.84AFLQDD149 pKa = 3.64LANEE153 pKa = 4.26YY154 pKa = 11.16GRR156 pKa = 11.84FLPALVMAHH165 pKa = 6.3EE166 pKa = 5.34FGHH169 pKa = 7.21AIQGRR174 pKa = 11.84VGYY177 pKa = 8.71PLQASIAIEE186 pKa = 4.03TQADD190 pKa = 3.98CFAGAWTAWVADD202 pKa = 3.86GNAPHH207 pKa = 5.84TTIRR211 pKa = 11.84TPEE214 pKa = 4.36LDD216 pKa = 3.62DD217 pKa = 3.56VLRR220 pKa = 11.84GYY222 pKa = 11.18LLLRR226 pKa = 11.84DD227 pKa = 4.03PVGTDD232 pKa = 3.31LNEE235 pKa = 4.38SEE237 pKa = 4.52AHH239 pKa = 6.15GSLFDD244 pKa = 3.77RR245 pKa = 11.84VSAFQQGFDD254 pKa = 4.79DD255 pKa = 5.89GPTACRR261 pKa = 11.84DD262 pKa = 3.22EE263 pKa = 4.9FGADD267 pKa = 3.14RR268 pKa = 11.84IYY270 pKa = 10.04TQRR273 pKa = 11.84EE274 pKa = 3.83FDD276 pKa = 3.84PADD279 pKa = 3.5AATQGNASYY288 pKa = 10.3PVTQQLVTEE297 pKa = 4.34SLSTFWSRR305 pKa = 11.84TFTARR310 pKa = 11.84GEE312 pKa = 4.22TFAAPDD318 pKa = 3.55VVPFSGTAPDD328 pKa = 4.45CAGQGNEE335 pKa = 3.82QADD338 pKa = 4.13LVFCANKK345 pKa = 9.2DD346 pKa = 3.52TVGYY350 pKa = 10.87DD351 pKa = 3.07EE352 pKa = 5.02TDD354 pKa = 3.1LVRR357 pKa = 11.84PVYY360 pKa = 10.55EE361 pKa = 4.04EE362 pKa = 4.33LGDD365 pKa = 3.95YY366 pKa = 10.99AVLTAVAIPYY376 pKa = 10.18SLAARR381 pKa = 11.84DD382 pKa = 3.82QLGLSTDD389 pKa = 3.71DD390 pKa = 3.14QDD392 pKa = 3.89AARR395 pKa = 11.84SAVCLTGAFTRR406 pKa = 11.84SVLSGEE412 pKa = 4.4VNGITISPGDD422 pKa = 3.81ADD424 pKa = 3.82EE425 pKa = 4.98SVQFLLGYY433 pKa = 10.29SDD435 pKa = 5.43DD436 pKa = 4.43PDD438 pKa = 3.59VLGDD442 pKa = 3.5VGLSGFQLVDD452 pKa = 3.04VFRR455 pKa = 11.84SGVVEE460 pKa = 4.36GEE462 pKa = 4.31SSCGIGG468 pKa = 3.84
Molecular weight: 49.81 kDa
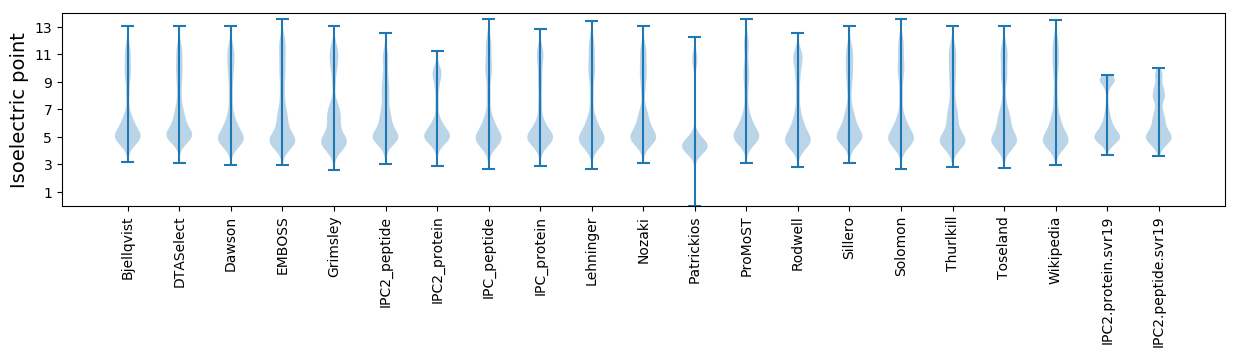
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H3LTL8|A0A1H3LTL8_9ACTN Endoglucanase OS=Modestobacter sp. DSM 44400 OX=1550230 GN=SAMN05661080_04249 PE=3 SV=1
MM1 pKa = 7.63RR2 pKa = 11.84RR3 pKa = 11.84TPAGAVPAVRR13 pKa = 11.84PVPVPGRR20 pKa = 11.84RR21 pKa = 11.84PPSRR25 pKa = 11.84SRR27 pKa = 11.84PPRR30 pKa = 11.84SPRR33 pKa = 11.84LRR35 pKa = 11.84SRR37 pKa = 11.84RR38 pKa = 11.84PRR40 pKa = 11.84SPRR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84SPRR48 pKa = 11.84RR49 pKa = 11.84PRR51 pKa = 11.84LRR53 pKa = 11.84RR54 pKa = 11.84LRR56 pKa = 11.84RR57 pKa = 11.84WLPRR61 pKa = 11.84PIVQPHH67 pKa = 6.04RR68 pKa = 11.84ARR70 pKa = 11.84ARR72 pKa = 11.84GRR74 pKa = 11.84VRR76 pKa = 11.84PPRR79 pKa = 11.84HH80 pKa = 6.06RR81 pKa = 11.84PPPSRR86 pKa = 11.84SRR88 pKa = 11.84LPSSRR93 pKa = 11.84LCSSRR98 pKa = 11.84RR99 pKa = 11.84PARR102 pKa = 11.84LSSSPRR108 pKa = 11.84PRR110 pKa = 11.84LSRR113 pKa = 11.84VRR115 pKa = 11.84VRR117 pKa = 11.84SGRR120 pKa = 11.84VRR122 pKa = 11.84PGAPPPVLRR131 pKa = 11.84VRR133 pKa = 11.84LVPVRR138 pKa = 11.84VPAPGRR144 pKa = 11.84RR145 pKa = 11.84RR146 pKa = 11.84VRR148 pKa = 11.84ATTPSAVPRR157 pKa = 11.84VRR159 pKa = 11.84PRR161 pKa = 11.84RR162 pKa = 11.84AARR165 pKa = 11.84LLPVRR170 pKa = 11.84ALPVRR175 pKa = 11.84ALPVRR180 pKa = 11.84ALPVRR185 pKa = 11.84ALPVRR190 pKa = 11.84VRR192 pKa = 11.84LARR195 pKa = 11.84AVPVPARR202 pKa = 11.84VPVDD206 pKa = 3.45LVRR209 pKa = 11.84RR210 pKa = 11.84PGPVDD215 pKa = 3.44LVRR218 pKa = 11.84RR219 pKa = 11.84PGPVDD224 pKa = 3.81LVPRR228 pKa = 11.84PAPVDD233 pKa = 3.59LVLPPVRR240 pKa = 11.84VGPGPTRR247 pKa = 11.84GG248 pKa = 3.31
MM1 pKa = 7.63RR2 pKa = 11.84RR3 pKa = 11.84TPAGAVPAVRR13 pKa = 11.84PVPVPGRR20 pKa = 11.84RR21 pKa = 11.84PPSRR25 pKa = 11.84SRR27 pKa = 11.84PPRR30 pKa = 11.84SPRR33 pKa = 11.84LRR35 pKa = 11.84SRR37 pKa = 11.84RR38 pKa = 11.84PRR40 pKa = 11.84SPRR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84SPRR48 pKa = 11.84RR49 pKa = 11.84PRR51 pKa = 11.84LRR53 pKa = 11.84RR54 pKa = 11.84LRR56 pKa = 11.84RR57 pKa = 11.84WLPRR61 pKa = 11.84PIVQPHH67 pKa = 6.04RR68 pKa = 11.84ARR70 pKa = 11.84ARR72 pKa = 11.84GRR74 pKa = 11.84VRR76 pKa = 11.84PPRR79 pKa = 11.84HH80 pKa = 6.06RR81 pKa = 11.84PPPSRR86 pKa = 11.84SRR88 pKa = 11.84LPSSRR93 pKa = 11.84LCSSRR98 pKa = 11.84RR99 pKa = 11.84PARR102 pKa = 11.84LSSSPRR108 pKa = 11.84PRR110 pKa = 11.84LSRR113 pKa = 11.84VRR115 pKa = 11.84VRR117 pKa = 11.84SGRR120 pKa = 11.84VRR122 pKa = 11.84PGAPPPVLRR131 pKa = 11.84VRR133 pKa = 11.84LVPVRR138 pKa = 11.84VPAPGRR144 pKa = 11.84RR145 pKa = 11.84RR146 pKa = 11.84VRR148 pKa = 11.84ATTPSAVPRR157 pKa = 11.84VRR159 pKa = 11.84PRR161 pKa = 11.84RR162 pKa = 11.84AARR165 pKa = 11.84LLPVRR170 pKa = 11.84ALPVRR175 pKa = 11.84ALPVRR180 pKa = 11.84ALPVRR185 pKa = 11.84ALPVRR190 pKa = 11.84VRR192 pKa = 11.84LARR195 pKa = 11.84AVPVPARR202 pKa = 11.84VPVDD206 pKa = 3.45LVRR209 pKa = 11.84RR210 pKa = 11.84PGPVDD215 pKa = 3.44LVRR218 pKa = 11.84RR219 pKa = 11.84PGPVDD224 pKa = 3.81LVPRR228 pKa = 11.84PAPVDD233 pKa = 3.59LVLPPVRR240 pKa = 11.84VGPGPTRR247 pKa = 11.84GG248 pKa = 3.31
Molecular weight: 27.92 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1452836 |
39 |
2006 |
282.7 |
29.96 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.334 ± 0.053 | 0.745 ± 0.011 |
6.144 ± 0.032 | 5.159 ± 0.029 |
2.533 ± 0.018 | 9.65 ± 0.034 |
2.078 ± 0.017 | 2.858 ± 0.025 |
1.439 ± 0.025 | 10.629 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.666 ± 0.012 | 1.516 ± 0.017 |
6.179 ± 0.032 | 2.856 ± 0.02 |
8.077 ± 0.036 | 5.156 ± 0.029 |
6.128 ± 0.026 | 9.603 ± 0.039 |
1.471 ± 0.014 | 1.779 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
