
Myceligenerans xiligouense
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Promicromonosporaceae; Myceligenerans
Average proteome isoelectric point is 6.01
Get precalculated fractions of proteins
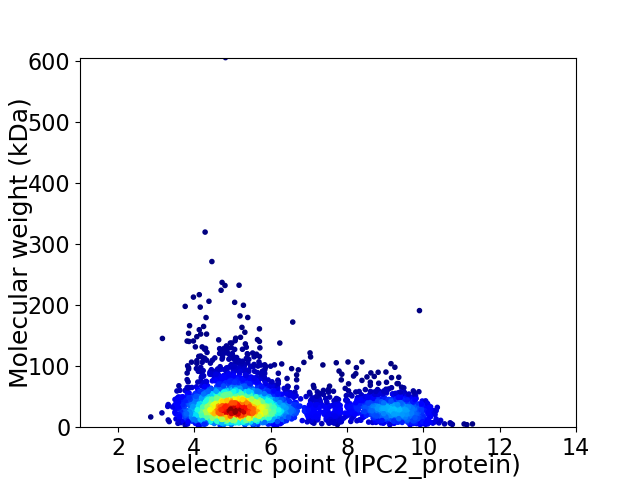
Virtual 2D-PAGE plot for 3975 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3N4ZCG6|A0A3N4ZCG6_9MICO Type II secretion system (T2SS) protein F OS=Myceligenerans xiligouense OX=253184 GN=EDD34_3843 PE=4 SV=1
MM1 pKa = 6.54QTRR4 pKa = 11.84KK5 pKa = 9.15QRR7 pKa = 11.84GRR9 pKa = 11.84PMAAAATALALAGVVLSPVTAQAEE33 pKa = 4.41DD34 pKa = 3.78GTEE37 pKa = 3.99LLTNPTFDD45 pKa = 3.69GTLDD49 pKa = 3.53PWWASEE55 pKa = 3.93GVEE58 pKa = 4.31FTEE61 pKa = 5.62APDD64 pKa = 3.61GTLCGVVPGGGNQWDD79 pKa = 4.29VIVGQGGVPVTAGDD93 pKa = 3.7TFTISVTAAASPEE106 pKa = 3.5ASARR110 pKa = 11.84VMVPDD115 pKa = 5.73PEE117 pKa = 4.73TEE119 pKa = 4.01WPPLYY124 pKa = 10.7YY125 pKa = 10.66ADD127 pKa = 3.2ITLRR131 pKa = 11.84ADD133 pKa = 3.29GQTFTEE139 pKa = 4.67AFDD142 pKa = 3.45IAADD146 pKa = 3.8AEE148 pKa = 4.84SEE150 pKa = 4.36VQLQLGGNAEE160 pKa = 4.15DD161 pKa = 5.85FEE163 pKa = 5.45LCLDD167 pKa = 4.0EE168 pKa = 5.89VSLLTGGEE176 pKa = 4.1VEE178 pKa = 4.41EE179 pKa = 4.86YY180 pKa = 10.48EE181 pKa = 4.57PEE183 pKa = 3.78TGPRR187 pKa = 11.84VRR189 pKa = 11.84VNQVGYY195 pKa = 10.72LPDD198 pKa = 3.93GPKK201 pKa = 10.39NATLVTDD208 pKa = 3.93ATSPVAWEE216 pKa = 4.49LRR218 pKa = 11.84DD219 pKa = 3.68SAGTAVASGEE229 pKa = 4.29TTPEE233 pKa = 3.89GTDD236 pKa = 3.23PSAGLNVHH244 pKa = 5.89TVDD247 pKa = 4.73FSGVDD252 pKa = 3.22ATGDD256 pKa = 3.91GFVLAADD263 pKa = 4.44GEE265 pKa = 4.31EE266 pKa = 4.44SYY268 pKa = 10.88PFEE271 pKa = 5.51ISATIYY277 pKa = 10.49DD278 pKa = 3.86QLRR281 pKa = 11.84ADD283 pKa = 3.87SLGIYY288 pKa = 7.12YY289 pKa = 7.43TQRR292 pKa = 11.84SGQEE296 pKa = 3.69IVPVEE301 pKa = 3.96VDD303 pKa = 3.53GEE305 pKa = 4.29QLKK308 pKa = 9.15TEE310 pKa = 4.2YY311 pKa = 10.55ARR313 pKa = 11.84PAGHH317 pKa = 6.2VSEE320 pKa = 5.73AGGDD324 pKa = 3.7DD325 pKa = 3.91VNQGDD330 pKa = 4.31VNVPCLPPEE339 pKa = 4.25GAVDD343 pKa = 3.54NSGNAQLGGDD353 pKa = 3.71DD354 pKa = 4.5HH355 pKa = 8.67YY356 pKa = 12.01GEE358 pKa = 5.23NGWACPEE365 pKa = 4.65GYY367 pKa = 10.61AIDD370 pKa = 5.23ASGGWYY376 pKa = 9.95DD377 pKa = 4.57AGDD380 pKa = 3.49HH381 pKa = 6.07GKK383 pKa = 10.29YY384 pKa = 9.76VVNSGISVYY393 pKa = 10.52QLLSTYY399 pKa = 9.53EE400 pKa = 3.75RR401 pKa = 11.84SLYY404 pKa = 10.83ADD406 pKa = 3.69TVSDD410 pKa = 3.99GALGDD415 pKa = 3.8STLVIPEE422 pKa = 4.3RR423 pKa = 11.84DD424 pKa = 3.3NGVPDD429 pKa = 3.43VLDD432 pKa = 3.87EE433 pKa = 4.34VRR435 pKa = 11.84HH436 pKa = 5.41NLDD439 pKa = 2.76WMLSMQVDD447 pKa = 4.32DD448 pKa = 4.59GVEE451 pKa = 3.76MTIDD455 pKa = 3.86GEE457 pKa = 4.34QYY459 pKa = 10.61DD460 pKa = 4.13AGGLAHH466 pKa = 7.08HH467 pKa = 7.05KK468 pKa = 10.21LHH470 pKa = 7.26DD471 pKa = 4.07IAWTGMNLLPHH482 pKa = 7.32EE483 pKa = 5.21DD484 pKa = 3.31PMPRR488 pKa = 11.84YY489 pKa = 8.6VHH491 pKa = 6.65RR492 pKa = 11.84PSTAATLNLAAAAAQGARR510 pKa = 11.84LYY512 pKa = 10.97ADD514 pKa = 4.3HH515 pKa = 7.26DD516 pKa = 3.54QAYY519 pKa = 10.59ADD521 pKa = 4.46EE522 pKa = 5.02LLDD525 pKa = 3.75AARR528 pKa = 11.84RR529 pKa = 11.84AWDD532 pKa = 3.5AASAHH537 pKa = 6.98PDD539 pKa = 3.32IIAPDD544 pKa = 4.07TNSLDD549 pKa = 3.93PNPGGGPYY557 pKa = 9.93NDD559 pKa = 3.9SSVGDD564 pKa = 3.69EE565 pKa = 5.01FYY567 pKa = 10.44WAAAQLFLTTGEE579 pKa = 4.38TEE581 pKa = 3.96FADD584 pKa = 3.94AVQSSPYY591 pKa = 9.81HH592 pKa = 5.95VGGAEE597 pKa = 3.87GDD599 pKa = 2.85IWRR602 pKa = 11.84ATGFDD607 pKa = 3.2WGFTAAAARR616 pKa = 11.84LDD618 pKa = 3.95LATVPNALPGRR629 pKa = 11.84DD630 pKa = 3.21AVVDD634 pKa = 4.14SVIEE638 pKa = 4.18GADD641 pKa = 3.14RR642 pKa = 11.84HH643 pKa = 5.97VATQQDD649 pKa = 4.19EE650 pKa = 4.85PFGHH654 pKa = 7.28AYY656 pKa = 10.45DD657 pKa = 4.78PGDD660 pKa = 3.73YY661 pKa = 10.59DD662 pKa = 3.02WGSNHH667 pKa = 5.91QVINNAVIIATAYY680 pKa = 10.67DD681 pKa = 3.75LTGEE685 pKa = 4.03AQYY688 pKa = 11.29RR689 pKa = 11.84DD690 pKa = 3.52AALEE694 pKa = 4.02TFDD697 pKa = 4.72YY698 pKa = 11.2VLGRR702 pKa = 11.84NALNNSYY709 pKa = 10.96VKK711 pKa = 10.79GYY713 pKa = 7.92GTQFSQNMHH722 pKa = 5.83NRR724 pKa = 11.84WMASADD730 pKa = 4.05RR731 pKa = 11.84LPPHH735 pKa = 7.4PDD737 pKa = 2.59GMLAAGPNSAIQDD750 pKa = 4.15PIAQEE755 pKa = 4.1KK756 pKa = 10.07LQGCAPQFCYY766 pKa = 9.79IDD768 pKa = 4.97EE769 pKa = 4.31VDD771 pKa = 3.25SWSTNEE777 pKa = 4.19MTINWNAPLSWFASWADD794 pKa = 3.59DD795 pKa = 3.58VANAGGDD802 pKa = 3.95VIADD806 pKa = 3.46ACTAEE811 pKa = 4.46LTVHH815 pKa = 6.47SDD817 pKa = 2.69WGSGFHH823 pKa = 5.62ATVRR827 pKa = 11.84VTAGDD832 pKa = 3.82APLSGWTTSFDD843 pKa = 3.55LGQATLANGWNAEE856 pKa = 3.93FSTSGSQVTASDD868 pKa = 3.82AGWNGTMAEE877 pKa = 4.26GGSTEE882 pKa = 4.02FGFVGSGAPPSSIPVACSAGG902 pKa = 3.39
MM1 pKa = 6.54QTRR4 pKa = 11.84KK5 pKa = 9.15QRR7 pKa = 11.84GRR9 pKa = 11.84PMAAAATALALAGVVLSPVTAQAEE33 pKa = 4.41DD34 pKa = 3.78GTEE37 pKa = 3.99LLTNPTFDD45 pKa = 3.69GTLDD49 pKa = 3.53PWWASEE55 pKa = 3.93GVEE58 pKa = 4.31FTEE61 pKa = 5.62APDD64 pKa = 3.61GTLCGVVPGGGNQWDD79 pKa = 4.29VIVGQGGVPVTAGDD93 pKa = 3.7TFTISVTAAASPEE106 pKa = 3.5ASARR110 pKa = 11.84VMVPDD115 pKa = 5.73PEE117 pKa = 4.73TEE119 pKa = 4.01WPPLYY124 pKa = 10.7YY125 pKa = 10.66ADD127 pKa = 3.2ITLRR131 pKa = 11.84ADD133 pKa = 3.29GQTFTEE139 pKa = 4.67AFDD142 pKa = 3.45IAADD146 pKa = 3.8AEE148 pKa = 4.84SEE150 pKa = 4.36VQLQLGGNAEE160 pKa = 4.15DD161 pKa = 5.85FEE163 pKa = 5.45LCLDD167 pKa = 4.0EE168 pKa = 5.89VSLLTGGEE176 pKa = 4.1VEE178 pKa = 4.41EE179 pKa = 4.86YY180 pKa = 10.48EE181 pKa = 4.57PEE183 pKa = 3.78TGPRR187 pKa = 11.84VRR189 pKa = 11.84VNQVGYY195 pKa = 10.72LPDD198 pKa = 3.93GPKK201 pKa = 10.39NATLVTDD208 pKa = 3.93ATSPVAWEE216 pKa = 4.49LRR218 pKa = 11.84DD219 pKa = 3.68SAGTAVASGEE229 pKa = 4.29TTPEE233 pKa = 3.89GTDD236 pKa = 3.23PSAGLNVHH244 pKa = 5.89TVDD247 pKa = 4.73FSGVDD252 pKa = 3.22ATGDD256 pKa = 3.91GFVLAADD263 pKa = 4.44GEE265 pKa = 4.31EE266 pKa = 4.44SYY268 pKa = 10.88PFEE271 pKa = 5.51ISATIYY277 pKa = 10.49DD278 pKa = 3.86QLRR281 pKa = 11.84ADD283 pKa = 3.87SLGIYY288 pKa = 7.12YY289 pKa = 7.43TQRR292 pKa = 11.84SGQEE296 pKa = 3.69IVPVEE301 pKa = 3.96VDD303 pKa = 3.53GEE305 pKa = 4.29QLKK308 pKa = 9.15TEE310 pKa = 4.2YY311 pKa = 10.55ARR313 pKa = 11.84PAGHH317 pKa = 6.2VSEE320 pKa = 5.73AGGDD324 pKa = 3.7DD325 pKa = 3.91VNQGDD330 pKa = 4.31VNVPCLPPEE339 pKa = 4.25GAVDD343 pKa = 3.54NSGNAQLGGDD353 pKa = 3.71DD354 pKa = 4.5HH355 pKa = 8.67YY356 pKa = 12.01GEE358 pKa = 5.23NGWACPEE365 pKa = 4.65GYY367 pKa = 10.61AIDD370 pKa = 5.23ASGGWYY376 pKa = 9.95DD377 pKa = 4.57AGDD380 pKa = 3.49HH381 pKa = 6.07GKK383 pKa = 10.29YY384 pKa = 9.76VVNSGISVYY393 pKa = 10.52QLLSTYY399 pKa = 9.53EE400 pKa = 3.75RR401 pKa = 11.84SLYY404 pKa = 10.83ADD406 pKa = 3.69TVSDD410 pKa = 3.99GALGDD415 pKa = 3.8STLVIPEE422 pKa = 4.3RR423 pKa = 11.84DD424 pKa = 3.3NGVPDD429 pKa = 3.43VLDD432 pKa = 3.87EE433 pKa = 4.34VRR435 pKa = 11.84HH436 pKa = 5.41NLDD439 pKa = 2.76WMLSMQVDD447 pKa = 4.32DD448 pKa = 4.59GVEE451 pKa = 3.76MTIDD455 pKa = 3.86GEE457 pKa = 4.34QYY459 pKa = 10.61DD460 pKa = 4.13AGGLAHH466 pKa = 7.08HH467 pKa = 7.05KK468 pKa = 10.21LHH470 pKa = 7.26DD471 pKa = 4.07IAWTGMNLLPHH482 pKa = 7.32EE483 pKa = 5.21DD484 pKa = 3.31PMPRR488 pKa = 11.84YY489 pKa = 8.6VHH491 pKa = 6.65RR492 pKa = 11.84PSTAATLNLAAAAAQGARR510 pKa = 11.84LYY512 pKa = 10.97ADD514 pKa = 4.3HH515 pKa = 7.26DD516 pKa = 3.54QAYY519 pKa = 10.59ADD521 pKa = 4.46EE522 pKa = 5.02LLDD525 pKa = 3.75AARR528 pKa = 11.84RR529 pKa = 11.84AWDD532 pKa = 3.5AASAHH537 pKa = 6.98PDD539 pKa = 3.32IIAPDD544 pKa = 4.07TNSLDD549 pKa = 3.93PNPGGGPYY557 pKa = 9.93NDD559 pKa = 3.9SSVGDD564 pKa = 3.69EE565 pKa = 5.01FYY567 pKa = 10.44WAAAQLFLTTGEE579 pKa = 4.38TEE581 pKa = 3.96FADD584 pKa = 3.94AVQSSPYY591 pKa = 9.81HH592 pKa = 5.95VGGAEE597 pKa = 3.87GDD599 pKa = 2.85IWRR602 pKa = 11.84ATGFDD607 pKa = 3.2WGFTAAAARR616 pKa = 11.84LDD618 pKa = 3.95LATVPNALPGRR629 pKa = 11.84DD630 pKa = 3.21AVVDD634 pKa = 4.14SVIEE638 pKa = 4.18GADD641 pKa = 3.14RR642 pKa = 11.84HH643 pKa = 5.97VATQQDD649 pKa = 4.19EE650 pKa = 4.85PFGHH654 pKa = 7.28AYY656 pKa = 10.45DD657 pKa = 4.78PGDD660 pKa = 3.73YY661 pKa = 10.59DD662 pKa = 3.02WGSNHH667 pKa = 5.91QVINNAVIIATAYY680 pKa = 10.67DD681 pKa = 3.75LTGEE685 pKa = 4.03AQYY688 pKa = 11.29RR689 pKa = 11.84DD690 pKa = 3.52AALEE694 pKa = 4.02TFDD697 pKa = 4.72YY698 pKa = 11.2VLGRR702 pKa = 11.84NALNNSYY709 pKa = 10.96VKK711 pKa = 10.79GYY713 pKa = 7.92GTQFSQNMHH722 pKa = 5.83NRR724 pKa = 11.84WMASADD730 pKa = 4.05RR731 pKa = 11.84LPPHH735 pKa = 7.4PDD737 pKa = 2.59GMLAAGPNSAIQDD750 pKa = 4.15PIAQEE755 pKa = 4.1KK756 pKa = 10.07LQGCAPQFCYY766 pKa = 9.79IDD768 pKa = 4.97EE769 pKa = 4.31VDD771 pKa = 3.25SWSTNEE777 pKa = 4.19MTINWNAPLSWFASWADD794 pKa = 3.59DD795 pKa = 3.58VANAGGDD802 pKa = 3.95VIADD806 pKa = 3.46ACTAEE811 pKa = 4.46LTVHH815 pKa = 6.47SDD817 pKa = 2.69WGSGFHH823 pKa = 5.62ATVRR827 pKa = 11.84VTAGDD832 pKa = 3.82APLSGWTTSFDD843 pKa = 3.55LGQATLANGWNAEE856 pKa = 3.93FSTSGSQVTASDD868 pKa = 3.82AGWNGTMAEE877 pKa = 4.26GGSTEE882 pKa = 4.02FGFVGSGAPPSSIPVACSAGG902 pKa = 3.39
Molecular weight: 95.21 kDa
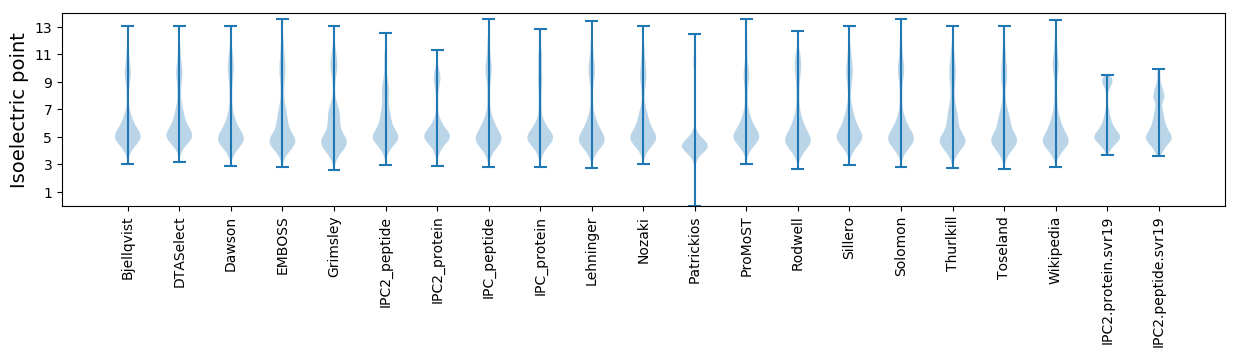
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3N4Z434|A0A3N4Z434_9MICO Uncharacterized protein OS=Myceligenerans xiligouense OX=253184 GN=EDD34_0524 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84TILANRR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.45GRR40 pKa = 11.84AKK42 pKa = 10.69LSAA45 pKa = 3.92
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84TILANRR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.45GRR40 pKa = 11.84AKK42 pKa = 10.69LSAA45 pKa = 3.92
Molecular weight: 5.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1408027 |
29 |
5751 |
354.2 |
37.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.769 ± 0.065 | 0.586 ± 0.009 |
6.649 ± 0.032 | 5.781 ± 0.038 |
2.643 ± 0.021 | 9.701 ± 0.036 |
2.147 ± 0.018 | 3.242 ± 0.026 |
1.649 ± 0.027 | 9.726 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.727 ± 0.016 | 1.757 ± 0.021 |
5.98 ± 0.034 | 2.545 ± 0.018 |
7.798 ± 0.047 | 5.133 ± 0.029 |
6.409 ± 0.032 | 9.233 ± 0.039 |
1.591 ± 0.019 | 1.932 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
