
Paracoccus tibetensis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Paracoccus
Average proteome isoelectric point is 6.46
Get precalculated fractions of proteins
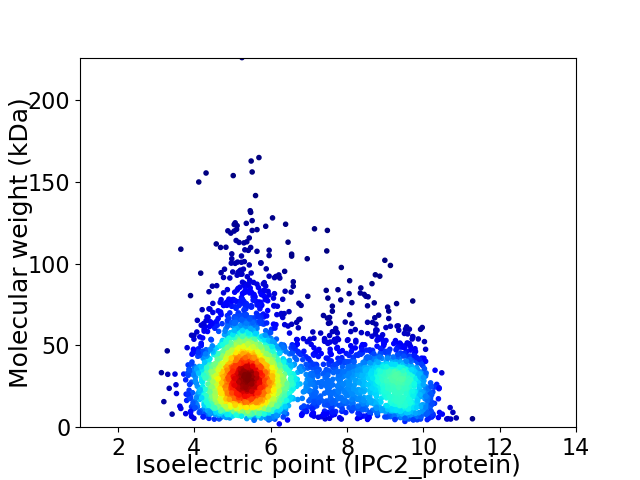
Virtual 2D-PAGE plot for 3733 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1G5HMT8|A0A1G5HMT8_9RHOB GTPase G3E family OS=Paracoccus tibetensis OX=336292 GN=SAMN05660710_02198 PE=3 SV=1
MM1 pKa = 7.85LEE3 pKa = 3.98RR4 pKa = 11.84LALATAVAAFPLMAYY19 pKa = 9.6AQTAEE24 pKa = 4.46TEE26 pKa = 4.35TPPDD30 pKa = 3.14AEE32 pKa = 4.43AAVEE36 pKa = 4.39TPVEE40 pKa = 4.07EE41 pKa = 4.47TPAADD46 pKa = 3.65AAATEE51 pKa = 4.21ATTDD55 pKa = 4.01DD56 pKa = 3.83VATDD60 pKa = 3.78DD61 pKa = 4.63EE62 pKa = 4.49ATEE65 pKa = 5.02DD66 pKa = 3.56EE67 pKa = 4.75AAPADD72 pKa = 4.25GSDD75 pKa = 3.13EE76 pKa = 3.97AAAEE80 pKa = 4.2GEE82 pKa = 4.31PAAEE86 pKa = 4.27AAAEE90 pKa = 4.23DD91 pKa = 4.08ADD93 pKa = 4.29AGAFTLPEE101 pKa = 4.13LGYY104 pKa = 10.71AYY106 pKa = 10.1DD107 pKa = 3.77ALEE110 pKa = 4.15PVIDD114 pKa = 4.01AQTMEE119 pKa = 4.3LHH121 pKa = 6.69HH122 pKa = 6.6SRR124 pKa = 11.84HH125 pKa = 5.7HH126 pKa = 4.77QTFVDD131 pKa = 3.75NLNDD135 pKa = 3.56AVEE138 pKa = 4.41NGVIPADD145 pKa = 3.56IEE147 pKa = 4.0IEE149 pKa = 4.32EE150 pKa = 4.33ILASASTYY158 pKa = 9.89PNQVRR163 pKa = 11.84NSAGGHH169 pKa = 4.73WNHH172 pKa = 5.66TFFWEE177 pKa = 3.77IMAPEE182 pKa = 4.7GEE184 pKa = 4.2RR185 pKa = 11.84GEE187 pKa = 4.22MSDD190 pKa = 4.25ALSEE194 pKa = 4.94AITAVYY200 pKa = 10.11GSEE203 pKa = 4.17EE204 pKa = 4.02EE205 pKa = 4.46FRR207 pKa = 11.84TAFNDD212 pKa = 2.97AGAARR217 pKa = 11.84FGSGWVWLIVNDD229 pKa = 4.12ANQLEE234 pKa = 4.88ITTTPNQDD242 pKa = 3.41NPLMDD247 pKa = 4.26AAEE250 pKa = 4.23VQGTPIIGNDD260 pKa = 3.27VWEE263 pKa = 4.08HH264 pKa = 6.11AYY266 pKa = 10.28YY267 pKa = 11.05LNYY270 pKa = 9.41QNRR273 pKa = 11.84RR274 pKa = 11.84AEE276 pKa = 4.0YY277 pKa = 9.4LQNWWDD283 pKa = 3.57VVNWDD288 pKa = 3.99EE289 pKa = 4.47VSARR293 pKa = 11.84YY294 pKa = 7.96EE295 pKa = 3.75AALAEE300 pKa = 4.3
MM1 pKa = 7.85LEE3 pKa = 3.98RR4 pKa = 11.84LALATAVAAFPLMAYY19 pKa = 9.6AQTAEE24 pKa = 4.46TEE26 pKa = 4.35TPPDD30 pKa = 3.14AEE32 pKa = 4.43AAVEE36 pKa = 4.39TPVEE40 pKa = 4.07EE41 pKa = 4.47TPAADD46 pKa = 3.65AAATEE51 pKa = 4.21ATTDD55 pKa = 4.01DD56 pKa = 3.83VATDD60 pKa = 3.78DD61 pKa = 4.63EE62 pKa = 4.49ATEE65 pKa = 5.02DD66 pKa = 3.56EE67 pKa = 4.75AAPADD72 pKa = 4.25GSDD75 pKa = 3.13EE76 pKa = 3.97AAAEE80 pKa = 4.2GEE82 pKa = 4.31PAAEE86 pKa = 4.27AAAEE90 pKa = 4.23DD91 pKa = 4.08ADD93 pKa = 4.29AGAFTLPEE101 pKa = 4.13LGYY104 pKa = 10.71AYY106 pKa = 10.1DD107 pKa = 3.77ALEE110 pKa = 4.15PVIDD114 pKa = 4.01AQTMEE119 pKa = 4.3LHH121 pKa = 6.69HH122 pKa = 6.6SRR124 pKa = 11.84HH125 pKa = 5.7HH126 pKa = 4.77QTFVDD131 pKa = 3.75NLNDD135 pKa = 3.56AVEE138 pKa = 4.41NGVIPADD145 pKa = 3.56IEE147 pKa = 4.0IEE149 pKa = 4.32EE150 pKa = 4.33ILASASTYY158 pKa = 9.89PNQVRR163 pKa = 11.84NSAGGHH169 pKa = 4.73WNHH172 pKa = 5.66TFFWEE177 pKa = 3.77IMAPEE182 pKa = 4.7GEE184 pKa = 4.2RR185 pKa = 11.84GEE187 pKa = 4.22MSDD190 pKa = 4.25ALSEE194 pKa = 4.94AITAVYY200 pKa = 10.11GSEE203 pKa = 4.17EE204 pKa = 4.02EE205 pKa = 4.46FRR207 pKa = 11.84TAFNDD212 pKa = 2.97AGAARR217 pKa = 11.84FGSGWVWLIVNDD229 pKa = 4.12ANQLEE234 pKa = 4.88ITTTPNQDD242 pKa = 3.41NPLMDD247 pKa = 4.26AAEE250 pKa = 4.23VQGTPIIGNDD260 pKa = 3.27VWEE263 pKa = 4.08HH264 pKa = 6.11AYY266 pKa = 10.28YY267 pKa = 11.05LNYY270 pKa = 9.41QNRR273 pKa = 11.84RR274 pKa = 11.84AEE276 pKa = 4.0YY277 pKa = 9.4LQNWWDD283 pKa = 3.57VVNWDD288 pKa = 3.99EE289 pKa = 4.47VSARR293 pKa = 11.84YY294 pKa = 7.96EE295 pKa = 3.75AALAEE300 pKa = 4.3
Molecular weight: 32.5 kDa
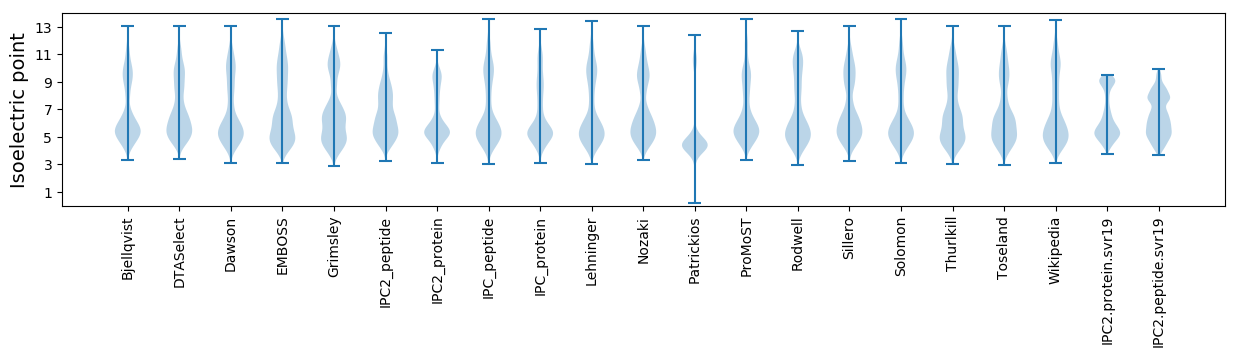
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1G5J786|A0A1G5J786_9RHOB 30S ribosomal protein S14 OS=Paracoccus tibetensis OX=336292 GN=rpsN PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.39GGRR28 pKa = 11.84LVLNRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.7GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.39GGRR28 pKa = 11.84LVLNRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.7GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1171507 |
18 |
2132 |
313.8 |
33.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.857 ± 0.069 | 0.808 ± 0.013 |
5.617 ± 0.032 | 5.761 ± 0.038 |
3.423 ± 0.025 | 8.89 ± 0.042 |
2.039 ± 0.022 | 4.94 ± 0.032 |
2.247 ± 0.031 | 10.636 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.74 ± 0.019 | 2.191 ± 0.021 |
5.609 ± 0.036 | 3.286 ± 0.027 |
7.681 ± 0.047 | 4.776 ± 0.027 |
5.098 ± 0.025 | 7.022 ± 0.034 |
1.458 ± 0.016 | 1.92 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
