
Dragonfly larvae associated circular virus-3
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 8.01
Get precalculated fractions of proteins
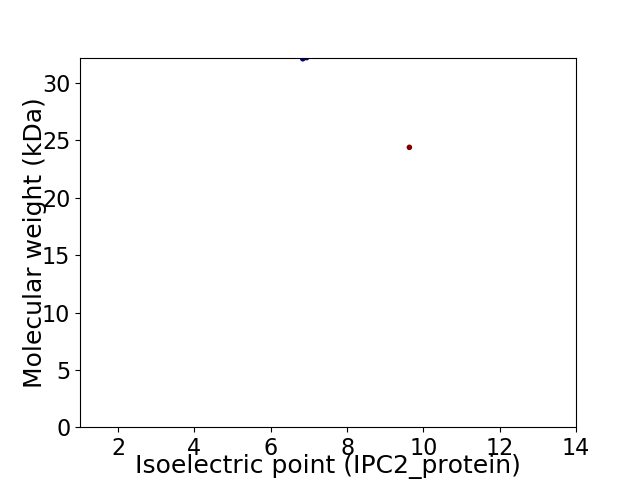
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W5U2P7|W5U2P7_9VIRU Replication-associated protein OS=Dragonfly larvae associated circular virus-3 OX=1454024 PE=4 SV=1
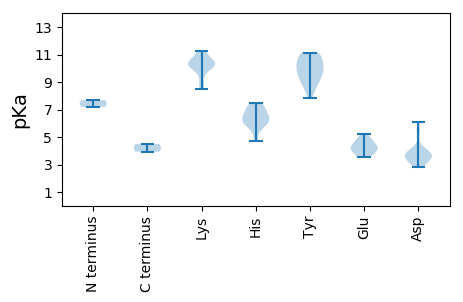
MM1 pKa = 7.69ANRR4 pKa = 11.84RR5 pKa = 11.84SRR7 pKa = 11.84GWCFTLNNYY16 pKa = 7.82TPEE19 pKa = 5.19HH20 pKa = 6.17EE21 pKa = 4.65ALLAAVPCAYY31 pKa = 9.85MIFGRR36 pKa = 11.84EE37 pKa = 3.71VGANGTPHH45 pKa = 6.6LQGFVYY51 pKa = 9.95FPNAKK56 pKa = 8.63TFNGAKK62 pKa = 9.88AVLPAGCHH70 pKa = 6.2LEE72 pKa = 3.81AAMGSVAQNVEE83 pKa = 4.3YY84 pKa = 10.41CSKK87 pKa = 11.09DD88 pKa = 2.85GDD90 pKa = 3.77VEE92 pKa = 4.31EE93 pKa = 5.04RR94 pKa = 11.84GEE96 pKa = 4.28RR97 pKa = 11.84PLSAAEE103 pKa = 4.24KK104 pKa = 10.0GASEE108 pKa = 3.97AQRR111 pKa = 11.84WKK113 pKa = 10.55DD114 pKa = 3.22ARR116 pKa = 11.84LAAVSGDD123 pKa = 3.35IADD126 pKa = 4.08VPDD129 pKa = 5.37DD130 pKa = 3.51IYY132 pKa = 10.94IRR134 pKa = 11.84YY135 pKa = 9.04YY136 pKa = 9.69RR137 pKa = 11.84TLKK140 pKa = 10.48EE141 pKa = 3.53ISKK144 pKa = 10.57DD145 pKa = 3.11HH146 pKa = 6.28MARR149 pKa = 11.84PDD151 pKa = 3.74GLDD154 pKa = 3.59GVCGLWLYY162 pKa = 10.93RR163 pKa = 11.84SAGTGKK169 pKa = 9.94SRR171 pKa = 11.84YY172 pKa = 8.88AQEE175 pKa = 4.2QFPEE179 pKa = 4.74HH180 pKa = 5.99YY181 pKa = 10.17MKK183 pKa = 8.5MTNKK187 pKa = 9.21WCDD190 pKa = 3.2GYY192 pKa = 11.06QGQDD196 pKa = 3.36VVVMDD201 pKa = 6.11DD202 pKa = 3.94MDD204 pKa = 5.44PDD206 pKa = 4.17HH207 pKa = 7.17ACLRR211 pKa = 11.84HH212 pKa = 5.75HH213 pKa = 7.46LKK215 pKa = 10.36RR216 pKa = 11.84WADD219 pKa = 3.79RR220 pKa = 11.84YY221 pKa = 10.38PFIGEE226 pKa = 4.28TKK228 pKa = 10.22GGAISIRR235 pKa = 11.84PKK237 pKa = 10.55KK238 pKa = 10.31FVVTSQYY245 pKa = 11.01SIEE248 pKa = 4.84DD249 pKa = 3.33MFKK252 pKa = 10.94NRR254 pKa = 11.84DD255 pKa = 3.08GTLDD259 pKa = 3.59VEE261 pKa = 4.72TVAAIRR267 pKa = 11.84RR268 pKa = 11.84RR269 pKa = 11.84FEE271 pKa = 3.87VKK273 pKa = 10.28RR274 pKa = 11.84FGVNAFNPYY283 pKa = 10.39NII285 pKa = 4.5
MM1 pKa = 7.69ANRR4 pKa = 11.84RR5 pKa = 11.84SRR7 pKa = 11.84GWCFTLNNYY16 pKa = 7.82TPEE19 pKa = 5.19HH20 pKa = 6.17EE21 pKa = 4.65ALLAAVPCAYY31 pKa = 9.85MIFGRR36 pKa = 11.84EE37 pKa = 3.71VGANGTPHH45 pKa = 6.6LQGFVYY51 pKa = 9.95FPNAKK56 pKa = 8.63TFNGAKK62 pKa = 9.88AVLPAGCHH70 pKa = 6.2LEE72 pKa = 3.81AAMGSVAQNVEE83 pKa = 4.3YY84 pKa = 10.41CSKK87 pKa = 11.09DD88 pKa = 2.85GDD90 pKa = 3.77VEE92 pKa = 4.31EE93 pKa = 5.04RR94 pKa = 11.84GEE96 pKa = 4.28RR97 pKa = 11.84PLSAAEE103 pKa = 4.24KK104 pKa = 10.0GASEE108 pKa = 3.97AQRR111 pKa = 11.84WKK113 pKa = 10.55DD114 pKa = 3.22ARR116 pKa = 11.84LAAVSGDD123 pKa = 3.35IADD126 pKa = 4.08VPDD129 pKa = 5.37DD130 pKa = 3.51IYY132 pKa = 10.94IRR134 pKa = 11.84YY135 pKa = 9.04YY136 pKa = 9.69RR137 pKa = 11.84TLKK140 pKa = 10.48EE141 pKa = 3.53ISKK144 pKa = 10.57DD145 pKa = 3.11HH146 pKa = 6.28MARR149 pKa = 11.84PDD151 pKa = 3.74GLDD154 pKa = 3.59GVCGLWLYY162 pKa = 10.93RR163 pKa = 11.84SAGTGKK169 pKa = 9.94SRR171 pKa = 11.84YY172 pKa = 8.88AQEE175 pKa = 4.2QFPEE179 pKa = 4.74HH180 pKa = 5.99YY181 pKa = 10.17MKK183 pKa = 8.5MTNKK187 pKa = 9.21WCDD190 pKa = 3.2GYY192 pKa = 11.06QGQDD196 pKa = 3.36VVVMDD201 pKa = 6.11DD202 pKa = 3.94MDD204 pKa = 5.44PDD206 pKa = 4.17HH207 pKa = 7.17ACLRR211 pKa = 11.84HH212 pKa = 5.75HH213 pKa = 7.46LKK215 pKa = 10.36RR216 pKa = 11.84WADD219 pKa = 3.79RR220 pKa = 11.84YY221 pKa = 10.38PFIGEE226 pKa = 4.28TKK228 pKa = 10.22GGAISIRR235 pKa = 11.84PKK237 pKa = 10.55KK238 pKa = 10.31FVVTSQYY245 pKa = 11.01SIEE248 pKa = 4.84DD249 pKa = 3.33MFKK252 pKa = 10.94NRR254 pKa = 11.84DD255 pKa = 3.08GTLDD259 pKa = 3.59VEE261 pKa = 4.72TVAAIRR267 pKa = 11.84RR268 pKa = 11.84RR269 pKa = 11.84FEE271 pKa = 3.87VKK273 pKa = 10.28RR274 pKa = 11.84FGVNAFNPYY283 pKa = 10.39NII285 pKa = 4.5
Molecular weight: 32.12 kDa
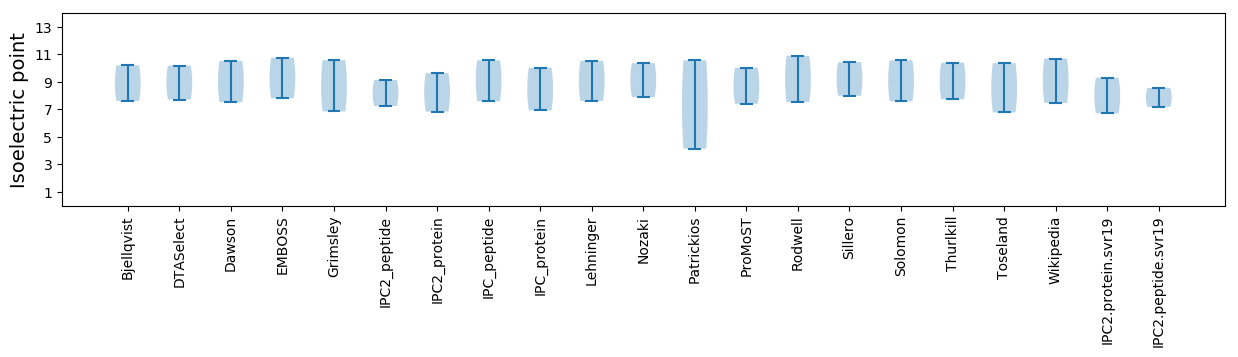
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W5U2P7|W5U2P7_9VIRU Replication-associated protein OS=Dragonfly larvae associated circular virus-3 OX=1454024 PE=4 SV=1
MM1 pKa = 7.2VLGKK5 pKa = 10.08RR6 pKa = 11.84GRR8 pKa = 11.84PGSMKK13 pKa = 10.27YY14 pKa = 9.62VPRR17 pKa = 11.84YY18 pKa = 9.15AKK20 pKa = 10.36RR21 pKa = 11.84ITRR24 pKa = 11.84GPYY27 pKa = 8.77VPRR30 pKa = 11.84GLTLRR35 pKa = 11.84PNNASVARR43 pKa = 11.84QVEE46 pKa = 4.56MKK48 pKa = 10.39FVDD51 pKa = 3.79ISPTFALDD59 pKa = 3.69FATPYY64 pKa = 10.43HH65 pKa = 6.63INLLNGIATGSDD77 pKa = 3.48LYY79 pKa = 11.14QRR81 pKa = 11.84VGRR84 pKa = 11.84SVYY87 pKa = 9.64MKK89 pKa = 10.2NLHH92 pKa = 7.22IDD94 pKa = 4.17FQCQNINTGSSTPDD108 pKa = 2.91LEE110 pKa = 4.18MRR112 pKa = 11.84VLVVLDD118 pKa = 3.78KK119 pKa = 11.27APNGAALPVLADD131 pKa = 3.9FLRR134 pKa = 11.84EE135 pKa = 3.69INAAGAATTNLNSHH149 pKa = 6.02QNTNNRR155 pKa = 11.84QRR157 pKa = 11.84FKK159 pKa = 11.29FLFDD163 pKa = 3.62KK164 pKa = 10.97KK165 pKa = 10.61IGIPPWNVGATTTGAPALNMVDD187 pKa = 3.78VYY189 pKa = 9.49WRR191 pKa = 11.84KK192 pKa = 9.81HH193 pKa = 4.72IKK195 pKa = 9.01CTTRR199 pKa = 11.84CSSLEE204 pKa = 3.93KK205 pKa = 10.46GHH207 pKa = 6.85RR208 pKa = 11.84CQKK211 pKa = 9.97SRR213 pKa = 11.84TMVYY217 pKa = 8.44TT218 pKa = 3.93
MM1 pKa = 7.2VLGKK5 pKa = 10.08RR6 pKa = 11.84GRR8 pKa = 11.84PGSMKK13 pKa = 10.27YY14 pKa = 9.62VPRR17 pKa = 11.84YY18 pKa = 9.15AKK20 pKa = 10.36RR21 pKa = 11.84ITRR24 pKa = 11.84GPYY27 pKa = 8.77VPRR30 pKa = 11.84GLTLRR35 pKa = 11.84PNNASVARR43 pKa = 11.84QVEE46 pKa = 4.56MKK48 pKa = 10.39FVDD51 pKa = 3.79ISPTFALDD59 pKa = 3.69FATPYY64 pKa = 10.43HH65 pKa = 6.63INLLNGIATGSDD77 pKa = 3.48LYY79 pKa = 11.14QRR81 pKa = 11.84VGRR84 pKa = 11.84SVYY87 pKa = 9.64MKK89 pKa = 10.2NLHH92 pKa = 7.22IDD94 pKa = 4.17FQCQNINTGSSTPDD108 pKa = 2.91LEE110 pKa = 4.18MRR112 pKa = 11.84VLVVLDD118 pKa = 3.78KK119 pKa = 11.27APNGAALPVLADD131 pKa = 3.9FLRR134 pKa = 11.84EE135 pKa = 3.69INAAGAATTNLNSHH149 pKa = 6.02QNTNNRR155 pKa = 11.84QRR157 pKa = 11.84FKK159 pKa = 11.29FLFDD163 pKa = 3.62KK164 pKa = 10.97KK165 pKa = 10.61IGIPPWNVGATTTGAPALNMVDD187 pKa = 3.78VYY189 pKa = 9.49WRR191 pKa = 11.84KK192 pKa = 9.81HH193 pKa = 4.72IKK195 pKa = 9.01CTTRR199 pKa = 11.84CSSLEE204 pKa = 3.93KK205 pKa = 10.46GHH207 pKa = 6.85RR208 pKa = 11.84CQKK211 pKa = 9.97SRR213 pKa = 11.84TMVYY217 pKa = 8.44TT218 pKa = 3.93
Molecular weight: 24.4 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
503 |
218 |
285 |
251.5 |
28.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.344 ± 1.036 | 2.187 ± 0.236 |
5.765 ± 1.097 | 4.175 ± 1.568 |
3.976 ± 0.205 | 7.753 ± 0.585 |
2.584 ± 0.195 | 4.175 ± 0.276 |
5.765 ± 0.133 | 6.561 ± 1.136 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.181 ± 0.02 | 5.567 ± 1.188 |
5.169 ± 0.532 | 2.982 ± 0.153 |
7.952 ± 0.204 | 4.573 ± 0.317 |
5.567 ± 1.495 | 6.958 ± 0.255 |
1.392 ± 0.318 | 4.374 ± 0.472 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
