
Halapricum sp. CBA1109
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Halobacteriales; Haloarculaceae; Halapricum; unclassified Halapricum
Average proteome isoelectric point is 4.92
Get precalculated fractions of proteins
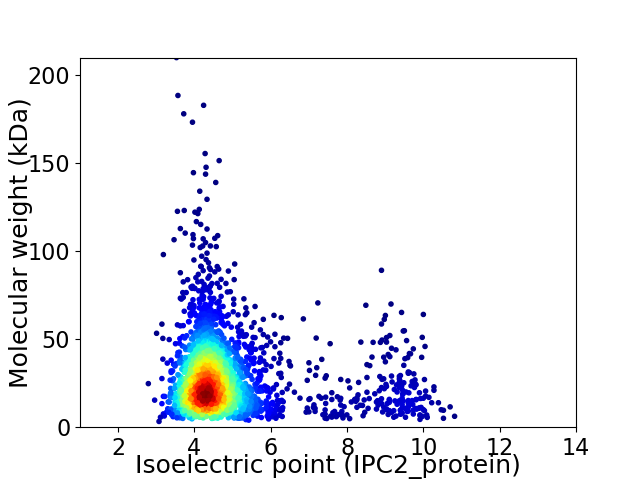
Virtual 2D-PAGE plot for 2712 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
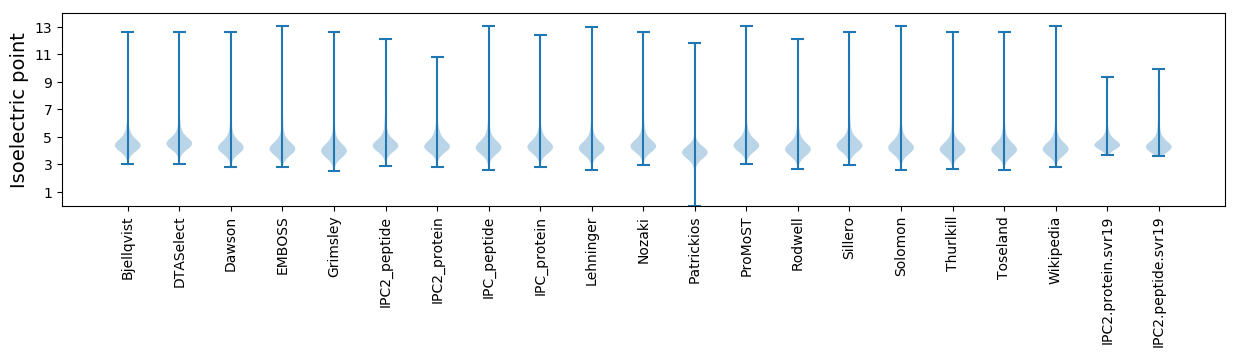
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6A9SZF5|A0A6A9SZF5_9EURY Uncharacterized protein OS=Halapricum sp. CBA1109 OX=2668068 GN=GJ629_09200 PE=4 SV=1
MM1 pKa = 7.58VDD3 pKa = 4.92SRR5 pKa = 11.84WDD7 pKa = 3.53DD8 pKa = 3.14QATYY12 pKa = 10.98YY13 pKa = 9.52NQTMPRR19 pKa = 11.84NCPPVTEE26 pKa = 5.61DD27 pKa = 3.88DD28 pKa = 3.81GTMDD32 pKa = 4.19GYY34 pKa = 10.83RR35 pKa = 11.84VEE37 pKa = 4.3GAIAGALYY45 pKa = 10.56DD46 pKa = 4.74VVDD49 pKa = 4.29DD50 pKa = 4.83TPNEE54 pKa = 4.02DD55 pKa = 3.53PMNEE59 pKa = 3.97SFATVYY65 pKa = 10.75SAFDD69 pKa = 3.32EE70 pKa = 4.46EE71 pKa = 4.88MPQSAFEE78 pKa = 4.47VYY80 pKa = 10.09DD81 pKa = 3.8ALAEE85 pKa = 4.11RR86 pKa = 11.84VDD88 pKa = 4.33NEE90 pKa = 4.08TALQQVFTTHH100 pKa = 7.19GIDD103 pKa = 3.38VAEE106 pKa = 4.66PALTVDD112 pKa = 4.36GGPNHH117 pKa = 6.68SSYY120 pKa = 11.77GNEE123 pKa = 3.81TTITGTAVDD132 pKa = 3.73EE133 pKa = 4.59HH134 pKa = 8.47SNITRR139 pKa = 11.84VQASVDD145 pKa = 3.43GGPWSTVAGDD155 pKa = 5.41DD156 pKa = 3.5ATEE159 pKa = 3.88WSVTVEE165 pKa = 4.29GPTDD169 pKa = 3.49TVTIRR174 pKa = 11.84ATDD177 pKa = 2.95HH178 pKa = 6.34WGNNATRR185 pKa = 11.84NVTVEE190 pKa = 3.88QSAVPSVVGGRR201 pKa = 11.84PITDD205 pKa = 3.54PDD207 pKa = 3.82GDD209 pKa = 4.02GVYY212 pKa = 10.74EE213 pKa = 4.54DD214 pKa = 4.02VDD216 pKa = 4.0GDD218 pKa = 4.16GEE220 pKa = 4.88ADD222 pKa = 3.71LFDD225 pKa = 4.62ALVYY229 pKa = 10.48YY230 pKa = 10.43NNRR233 pKa = 11.84NSGVIEE239 pKa = 3.83AHH241 pKa = 6.76PGLFDD246 pKa = 3.99YY247 pKa = 11.0DD248 pKa = 4.14SEE250 pKa = 5.82DD251 pKa = 4.14DD252 pKa = 3.84VGTLFDD258 pKa = 4.31ALALFEE264 pKa = 5.14MIGAPNRR271 pKa = 3.66
MM1 pKa = 7.58VDD3 pKa = 4.92SRR5 pKa = 11.84WDD7 pKa = 3.53DD8 pKa = 3.14QATYY12 pKa = 10.98YY13 pKa = 9.52NQTMPRR19 pKa = 11.84NCPPVTEE26 pKa = 5.61DD27 pKa = 3.88DD28 pKa = 3.81GTMDD32 pKa = 4.19GYY34 pKa = 10.83RR35 pKa = 11.84VEE37 pKa = 4.3GAIAGALYY45 pKa = 10.56DD46 pKa = 4.74VVDD49 pKa = 4.29DD50 pKa = 4.83TPNEE54 pKa = 4.02DD55 pKa = 3.53PMNEE59 pKa = 3.97SFATVYY65 pKa = 10.75SAFDD69 pKa = 3.32EE70 pKa = 4.46EE71 pKa = 4.88MPQSAFEE78 pKa = 4.47VYY80 pKa = 10.09DD81 pKa = 3.8ALAEE85 pKa = 4.11RR86 pKa = 11.84VDD88 pKa = 4.33NEE90 pKa = 4.08TALQQVFTTHH100 pKa = 7.19GIDD103 pKa = 3.38VAEE106 pKa = 4.66PALTVDD112 pKa = 4.36GGPNHH117 pKa = 6.68SSYY120 pKa = 11.77GNEE123 pKa = 3.81TTITGTAVDD132 pKa = 3.73EE133 pKa = 4.59HH134 pKa = 8.47SNITRR139 pKa = 11.84VQASVDD145 pKa = 3.43GGPWSTVAGDD155 pKa = 5.41DD156 pKa = 3.5ATEE159 pKa = 3.88WSVTVEE165 pKa = 4.29GPTDD169 pKa = 3.49TVTIRR174 pKa = 11.84ATDD177 pKa = 2.95HH178 pKa = 6.34WGNNATRR185 pKa = 11.84NVTVEE190 pKa = 3.88QSAVPSVVGGRR201 pKa = 11.84PITDD205 pKa = 3.54PDD207 pKa = 3.82GDD209 pKa = 4.02GVYY212 pKa = 10.74EE213 pKa = 4.54DD214 pKa = 4.02VDD216 pKa = 4.0GDD218 pKa = 4.16GEE220 pKa = 4.88ADD222 pKa = 3.71LFDD225 pKa = 4.62ALVYY229 pKa = 10.48YY230 pKa = 10.43NNRR233 pKa = 11.84NSGVIEE239 pKa = 3.83AHH241 pKa = 6.76PGLFDD246 pKa = 3.99YY247 pKa = 11.0DD248 pKa = 4.14SEE250 pKa = 5.82DD251 pKa = 4.14DD252 pKa = 3.84VGTLFDD258 pKa = 4.31ALALFEE264 pKa = 5.14MIGAPNRR271 pKa = 3.66
Molecular weight: 29.24 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6A9T8L7|A0A6A9T8L7_9EURY Uncharacterized protein OS=Halapricum sp. CBA1109 OX=2668068 GN=GJ629_05530 PE=4 SV=1
MM1 pKa = 7.28VVLLVVASAVPLARR15 pKa = 11.84PSPRR19 pKa = 11.84RR20 pKa = 11.84TAAVVTRR27 pKa = 11.84RR28 pKa = 11.84HH29 pKa = 4.96QPRR32 pKa = 11.84TTGRR36 pKa = 11.84RR37 pKa = 11.84HH38 pKa = 5.75PTTGPEE44 pKa = 3.46RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84PDD49 pKa = 2.86RR50 pKa = 11.84RR51 pKa = 11.84TRR53 pKa = 11.84RR54 pKa = 3.44
MM1 pKa = 7.28VVLLVVASAVPLARR15 pKa = 11.84PSPRR19 pKa = 11.84RR20 pKa = 11.84TAAVVTRR27 pKa = 11.84RR28 pKa = 11.84HH29 pKa = 4.96QPRR32 pKa = 11.84TTGRR36 pKa = 11.84RR37 pKa = 11.84HH38 pKa = 5.75PTTGPEE44 pKa = 3.46RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84PDD49 pKa = 2.86RR50 pKa = 11.84RR51 pKa = 11.84TRR53 pKa = 11.84RR54 pKa = 3.44
Molecular weight: 6.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
727707 |
31 |
1997 |
268.3 |
29.06 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.874 ± 0.063 | 0.688 ± 0.013 |
8.704 ± 0.065 | 8.032 ± 0.071 |
3.216 ± 0.035 | 8.696 ± 0.05 |
1.882 ± 0.027 | 3.871 ± 0.037 |
1.635 ± 0.03 | 8.653 ± 0.063 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.712 ± 0.022 | 2.243 ± 0.03 |
4.771 ± 0.038 | 2.528 ± 0.031 |
6.686 ± 0.049 | 5.79 ± 0.04 |
6.878 ± 0.052 | 9.276 ± 0.058 |
1.111 ± 0.019 | 2.755 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
