
Aspergillus nomiae NRRL 13137
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; Eurotiomycetes; Eurotiomycetidae; Eurotiales; Aspergillaceae; Aspergillus; Aspergillus nomiae
Average proteome isoelectric point is 6.33
Get precalculated fractions of proteins
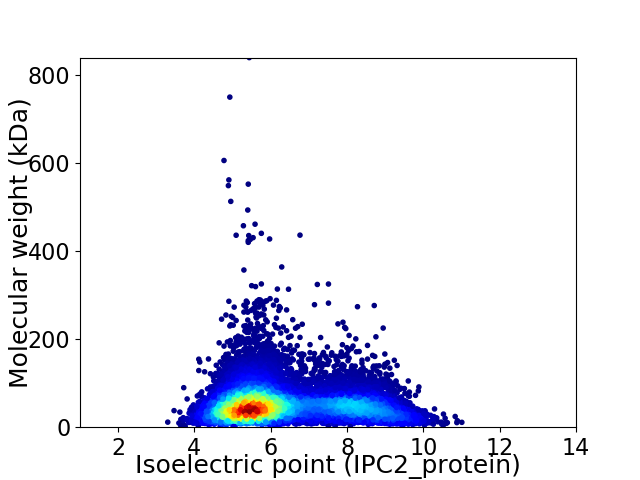
Virtual 2D-PAGE plot for 11914 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
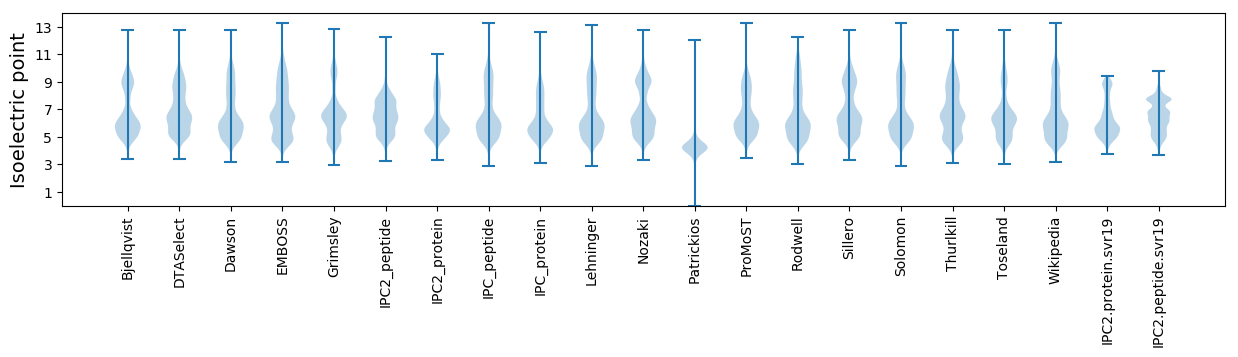
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0L1JGA0|A0A0L1JGA0_ASPNO Peroxisomal hydratase-dehydrogenase-epimerase OS=Aspergillus nomiae NRRL 13137 OX=1509407 GN=ANOM_001084 PE=4 SV=1
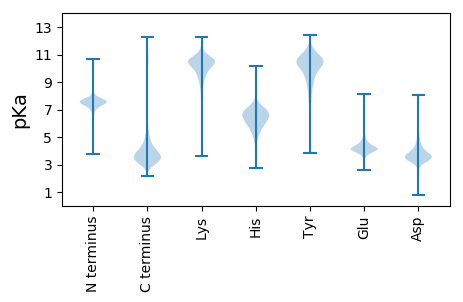
MM1 pKa = 7.83RR2 pKa = 11.84PYY4 pKa = 11.25SLLSSLAILGPAALTVAQTLNIVAHH29 pKa = 6.27QDD31 pKa = 3.27DD32 pKa = 5.27DD33 pKa = 4.76LLFMSPDD40 pKa = 3.31LLSEE44 pKa = 4.24VRR46 pKa = 11.84SGRR49 pKa = 11.84AVRR52 pKa = 11.84TVFLTAGDD60 pKa = 3.83AGNGEE65 pKa = 5.03DD66 pKa = 3.87YY67 pKa = 7.67WTSRR71 pKa = 11.84QAGSLATYY79 pKa = 9.88AQIAGVANEE88 pKa = 4.23WNEE91 pKa = 3.36GDD93 pKa = 3.81AGIEE97 pKa = 4.1GFDD100 pKa = 3.28IPVYY104 pKa = 10.26EE105 pKa = 5.35LAAQPQIEE113 pKa = 4.62LAFLHH118 pKa = 6.87IPDD121 pKa = 4.77GNLDD125 pKa = 3.51GSGFASTGSVSLQKK139 pKa = 10.6LWEE142 pKa = 4.16GTIDD146 pKa = 4.27QIGTVDD152 pKa = 3.93ASGTTYY158 pKa = 10.16TRR160 pKa = 11.84QQLLDD165 pKa = 3.66VLSDD169 pKa = 3.65IIEE172 pKa = 4.39NFSPDD177 pKa = 4.18RR178 pKa = 11.84INTLDD183 pKa = 3.59FVNDD187 pKa = 3.65IGDD190 pKa = 4.05GDD192 pKa = 4.26HH193 pKa = 7.15SDD195 pKa = 3.52HH196 pKa = 6.12YY197 pKa = 9.18TTGFFADD204 pKa = 4.37HH205 pKa = 7.14ASQAADD211 pKa = 3.22NNADD215 pKa = 3.27FFGYY219 pKa = 8.88MGYY222 pKa = 9.61PVASLPANLSPDD234 pKa = 3.44QIADD238 pKa = 3.44KK239 pKa = 10.91KK240 pKa = 11.12AIFYY244 pKa = 9.38FYY246 pKa = 10.9AGYY249 pKa = 10.77DD250 pKa = 3.43SGTCNSDD257 pKa = 3.21AACAGRR263 pKa = 11.84PEE265 pKa = 4.35LAWLEE270 pKa = 4.0RR271 pKa = 11.84QYY273 pKa = 11.48QVV275 pKa = 3.32
MM1 pKa = 7.83RR2 pKa = 11.84PYY4 pKa = 11.25SLLSSLAILGPAALTVAQTLNIVAHH29 pKa = 6.27QDD31 pKa = 3.27DD32 pKa = 5.27DD33 pKa = 4.76LLFMSPDD40 pKa = 3.31LLSEE44 pKa = 4.24VRR46 pKa = 11.84SGRR49 pKa = 11.84AVRR52 pKa = 11.84TVFLTAGDD60 pKa = 3.83AGNGEE65 pKa = 5.03DD66 pKa = 3.87YY67 pKa = 7.67WTSRR71 pKa = 11.84QAGSLATYY79 pKa = 9.88AQIAGVANEE88 pKa = 4.23WNEE91 pKa = 3.36GDD93 pKa = 3.81AGIEE97 pKa = 4.1GFDD100 pKa = 3.28IPVYY104 pKa = 10.26EE105 pKa = 5.35LAAQPQIEE113 pKa = 4.62LAFLHH118 pKa = 6.87IPDD121 pKa = 4.77GNLDD125 pKa = 3.51GSGFASTGSVSLQKK139 pKa = 10.6LWEE142 pKa = 4.16GTIDD146 pKa = 4.27QIGTVDD152 pKa = 3.93ASGTTYY158 pKa = 10.16TRR160 pKa = 11.84QQLLDD165 pKa = 3.66VLSDD169 pKa = 3.65IIEE172 pKa = 4.39NFSPDD177 pKa = 4.18RR178 pKa = 11.84INTLDD183 pKa = 3.59FVNDD187 pKa = 3.65IGDD190 pKa = 4.05GDD192 pKa = 4.26HH193 pKa = 7.15SDD195 pKa = 3.52HH196 pKa = 6.12YY197 pKa = 9.18TTGFFADD204 pKa = 4.37HH205 pKa = 7.14ASQAADD211 pKa = 3.22NNADD215 pKa = 3.27FFGYY219 pKa = 8.88MGYY222 pKa = 9.61PVASLPANLSPDD234 pKa = 3.44QIADD238 pKa = 3.44KK239 pKa = 10.91KK240 pKa = 11.12AIFYY244 pKa = 9.38FYY246 pKa = 10.9AGYY249 pKa = 10.77DD250 pKa = 3.43SGTCNSDD257 pKa = 3.21AACAGRR263 pKa = 11.84PEE265 pKa = 4.35LAWLEE270 pKa = 4.0RR271 pKa = 11.84QYY273 pKa = 11.48QVV275 pKa = 3.32
Molecular weight: 29.62 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0L1ILW8|A0A0L1ILW8_ASPNO Arginine permease OS=Aspergillus nomiae NRRL 13137 OX=1509407 GN=ANOM_011281 PE=4 SV=1
LL1 pKa = 7.45LLPSLLPPLLLPSLLPPLLRR21 pKa = 11.84LGLLLLLLRR30 pKa = 11.84LGLLLQRR37 pKa = 11.84RR38 pKa = 11.84LLSLLPLLRR47 pKa = 11.84PMFPPFSLPLVVPVFLASLLSPAPLLSRR75 pKa = 11.84LPPLRR80 pKa = 11.84PSPVSPLSSQLFQVLTLPLLPP101 pKa = 5.06
LL1 pKa = 7.45LLPSLLPPLLLPSLLPPLLRR21 pKa = 11.84LGLLLLLLRR30 pKa = 11.84LGLLLQRR37 pKa = 11.84RR38 pKa = 11.84LLSLLPLLRR47 pKa = 11.84PMFPPFSLPLVVPVFLASLLSPAPLLSRR75 pKa = 11.84LPPLRR80 pKa = 11.84PSPVSPLSSQLFQVLTLPLLPP101 pKa = 5.06
Molecular weight: 11.04 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5973996 |
11 |
7584 |
501.4 |
55.59 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.334 ± 0.018 | 1.311 ± 0.008 |
5.598 ± 0.015 | 6.053 ± 0.022 |
3.818 ± 0.014 | 6.794 ± 0.019 |
2.446 ± 0.008 | 5.113 ± 0.016 |
4.553 ± 0.018 | 9.242 ± 0.023 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.164 ± 0.008 | 3.679 ± 0.011 |
5.9 ± 0.024 | 4.033 ± 0.013 |
5.996 ± 0.018 | 8.268 ± 0.022 |
5.955 ± 0.013 | 6.294 ± 0.015 |
1.519 ± 0.009 | 2.93 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
