
Gramella fulva
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Gramella
Average proteome isoelectric point is 6.49
Get precalculated fractions of proteins
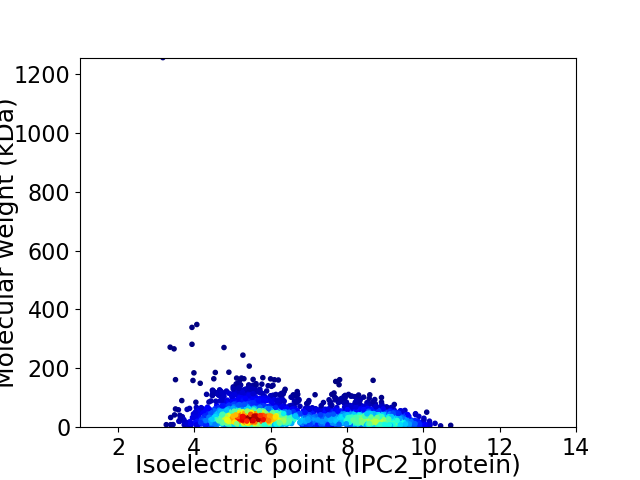
Virtual 2D-PAGE plot for 3785 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2R3ZB15|A0A2R3ZB15_9FLAO Probable potassium transport system protein kup OS=Gramella fulva OX=2126553 GN=trkD PE=3 SV=1
MM1 pKa = 7.33VKK3 pKa = 10.39GFLVAFQLLFFLMACSSEE21 pKa = 4.17DD22 pKa = 3.41TPGQPKK28 pKa = 8.11PTPDD32 pKa = 4.34PDD34 pKa = 3.67PVAKK38 pKa = 10.09ADD40 pKa = 3.67KK41 pKa = 10.63LSAVEE46 pKa = 3.93NEE48 pKa = 4.34EE49 pKa = 4.03YY50 pKa = 10.46TFQSSILLEE59 pKa = 4.04NDD61 pKa = 3.57EE62 pKa = 4.6IVDD65 pKa = 3.47NARR68 pKa = 11.84IKK70 pKa = 11.2DD71 pKa = 3.76FDD73 pKa = 3.98NEE75 pKa = 4.15TTAGGSITDD84 pKa = 4.08NRR86 pKa = 11.84DD87 pKa = 2.5GTYY90 pKa = 10.02TYY92 pKa = 9.91TPPADD97 pKa = 3.88FKK99 pKa = 11.73GDD101 pKa = 3.53DD102 pKa = 3.61TFDD105 pKa = 3.47YY106 pKa = 9.13TLCVPGDD113 pKa = 3.84SNRR116 pKa = 11.84CSTATVTISVGDD128 pKa = 3.7AGDD131 pKa = 3.57PVAIDD136 pKa = 4.19DD137 pKa = 4.47SYY139 pKa = 11.68QINEE143 pKa = 4.13NEE145 pKa = 4.38SYY147 pKa = 10.55TIRR150 pKa = 11.84NYY152 pKa = 11.22LNNDD156 pKa = 3.71DD157 pKa = 5.38LKK159 pKa = 11.67DD160 pKa = 3.65NATVTDD166 pKa = 3.8VLSEE170 pKa = 4.22SGNATVTLQEE180 pKa = 4.98DD181 pKa = 4.04GSIRR185 pKa = 11.84YY186 pKa = 8.56VPNEE190 pKa = 4.1HH191 pKa = 6.97FSGEE195 pKa = 4.09DD196 pKa = 2.98TFTYY200 pKa = 8.17TICDD204 pKa = 3.61DD205 pKa = 5.44DD206 pKa = 4.01EE207 pKa = 4.66TPNCSTATITMTVVDD222 pKa = 4.54EE223 pKa = 4.69GSPKK227 pKa = 10.79AEE229 pKa = 4.56DD230 pKa = 3.71DD231 pKa = 3.95SVIIASGTTEE241 pKa = 3.95TTFKK245 pKa = 11.25NLLDD249 pKa = 4.31NDD251 pKa = 4.5DD252 pKa = 5.55LIDD255 pKa = 4.23DD256 pKa = 4.12AVITSVEE263 pKa = 4.14STGNGTATLNEE274 pKa = 4.88DD275 pKa = 3.32GTVTFEE281 pKa = 4.05PQAGFSGDD289 pKa = 3.47DD290 pKa = 3.22TFTYY294 pKa = 8.15TICDD298 pKa = 3.65DD299 pKa = 5.88DD300 pKa = 4.63VDD302 pKa = 4.36QFCSTATVTVSVVEE316 pKa = 4.31SYY318 pKa = 11.33GFNIPADD325 pKa = 3.6LEE327 pKa = 4.45YY328 pKa = 10.86YY329 pKa = 10.58YY330 pKa = 11.44GDD332 pKa = 4.49AIFAKK337 pKa = 10.58GGGTIAYY344 pKa = 9.24QLLSDD349 pKa = 4.72FTNAMHH355 pKa = 6.05TTHH358 pKa = 7.36LEE360 pKa = 3.77YY361 pKa = 10.27TDD363 pKa = 3.42RR364 pKa = 11.84HH365 pKa = 6.45DD366 pKa = 3.97YY367 pKa = 11.13LYY369 pKa = 11.14DD370 pKa = 4.4ADD372 pKa = 5.65ADD374 pKa = 3.94PADD377 pKa = 4.1PSKK380 pKa = 11.17VILMYY385 pKa = 10.62SGEE388 pKa = 4.34SRR390 pKa = 11.84PDD392 pKa = 3.19DD393 pKa = 3.26QYY395 pKa = 12.02QLHH398 pKa = 6.44EE399 pKa = 4.53PPLRR403 pKa = 11.84GEE405 pKa = 4.39TFNTEE410 pKa = 3.91HH411 pKa = 7.62IYY413 pKa = 9.98PQSKK417 pKa = 9.96LDD419 pKa = 3.69HH420 pKa = 7.43DD421 pKa = 4.11EE422 pKa = 4.23AANDD426 pKa = 3.7LHH428 pKa = 7.69HH429 pKa = 6.62MRR431 pKa = 11.84VADD434 pKa = 3.17IDD436 pKa = 4.32VNSEE440 pKa = 3.26RR441 pKa = 11.84SYY443 pKa = 11.87YY444 pKa = 10.18PFTDD448 pKa = 3.32GTGTYY453 pKa = 10.8KK454 pKa = 10.87LVDD457 pKa = 3.63GNKK460 pKa = 8.95WFPGDD465 pKa = 3.52DD466 pKa = 3.11WRR468 pKa = 11.84GDD470 pKa = 3.45VARR473 pKa = 11.84MVMYY477 pKa = 11.02VNLAYY482 pKa = 10.23GDD484 pKa = 4.02SFDD487 pKa = 4.35EE488 pKa = 4.47VGSKK492 pKa = 10.92DD493 pKa = 6.14LFLKK497 pKa = 9.84WNRR500 pKa = 11.84EE501 pKa = 4.0DD502 pKa = 3.58PVSEE506 pKa = 5.39FEE508 pKa = 3.92IQRR511 pKa = 11.84NNVIEE516 pKa = 4.38GAQGNRR522 pKa = 11.84NPFIDD527 pKa = 3.96NPYY530 pKa = 10.19LATLIWGGEE539 pKa = 3.84PAQNRR544 pKa = 11.84WQQ546 pKa = 3.34
MM1 pKa = 7.33VKK3 pKa = 10.39GFLVAFQLLFFLMACSSEE21 pKa = 4.17DD22 pKa = 3.41TPGQPKK28 pKa = 8.11PTPDD32 pKa = 4.34PDD34 pKa = 3.67PVAKK38 pKa = 10.09ADD40 pKa = 3.67KK41 pKa = 10.63LSAVEE46 pKa = 3.93NEE48 pKa = 4.34EE49 pKa = 4.03YY50 pKa = 10.46TFQSSILLEE59 pKa = 4.04NDD61 pKa = 3.57EE62 pKa = 4.6IVDD65 pKa = 3.47NARR68 pKa = 11.84IKK70 pKa = 11.2DD71 pKa = 3.76FDD73 pKa = 3.98NEE75 pKa = 4.15TTAGGSITDD84 pKa = 4.08NRR86 pKa = 11.84DD87 pKa = 2.5GTYY90 pKa = 10.02TYY92 pKa = 9.91TPPADD97 pKa = 3.88FKK99 pKa = 11.73GDD101 pKa = 3.53DD102 pKa = 3.61TFDD105 pKa = 3.47YY106 pKa = 9.13TLCVPGDD113 pKa = 3.84SNRR116 pKa = 11.84CSTATVTISVGDD128 pKa = 3.7AGDD131 pKa = 3.57PVAIDD136 pKa = 4.19DD137 pKa = 4.47SYY139 pKa = 11.68QINEE143 pKa = 4.13NEE145 pKa = 4.38SYY147 pKa = 10.55TIRR150 pKa = 11.84NYY152 pKa = 11.22LNNDD156 pKa = 3.71DD157 pKa = 5.38LKK159 pKa = 11.67DD160 pKa = 3.65NATVTDD166 pKa = 3.8VLSEE170 pKa = 4.22SGNATVTLQEE180 pKa = 4.98DD181 pKa = 4.04GSIRR185 pKa = 11.84YY186 pKa = 8.56VPNEE190 pKa = 4.1HH191 pKa = 6.97FSGEE195 pKa = 4.09DD196 pKa = 2.98TFTYY200 pKa = 8.17TICDD204 pKa = 3.61DD205 pKa = 5.44DD206 pKa = 4.01EE207 pKa = 4.66TPNCSTATITMTVVDD222 pKa = 4.54EE223 pKa = 4.69GSPKK227 pKa = 10.79AEE229 pKa = 4.56DD230 pKa = 3.71DD231 pKa = 3.95SVIIASGTTEE241 pKa = 3.95TTFKK245 pKa = 11.25NLLDD249 pKa = 4.31NDD251 pKa = 4.5DD252 pKa = 5.55LIDD255 pKa = 4.23DD256 pKa = 4.12AVITSVEE263 pKa = 4.14STGNGTATLNEE274 pKa = 4.88DD275 pKa = 3.32GTVTFEE281 pKa = 4.05PQAGFSGDD289 pKa = 3.47DD290 pKa = 3.22TFTYY294 pKa = 8.15TICDD298 pKa = 3.65DD299 pKa = 5.88DD300 pKa = 4.63VDD302 pKa = 4.36QFCSTATVTVSVVEE316 pKa = 4.31SYY318 pKa = 11.33GFNIPADD325 pKa = 3.6LEE327 pKa = 4.45YY328 pKa = 10.86YY329 pKa = 10.58YY330 pKa = 11.44GDD332 pKa = 4.49AIFAKK337 pKa = 10.58GGGTIAYY344 pKa = 9.24QLLSDD349 pKa = 4.72FTNAMHH355 pKa = 6.05TTHH358 pKa = 7.36LEE360 pKa = 3.77YY361 pKa = 10.27TDD363 pKa = 3.42RR364 pKa = 11.84HH365 pKa = 6.45DD366 pKa = 3.97YY367 pKa = 11.13LYY369 pKa = 11.14DD370 pKa = 4.4ADD372 pKa = 5.65ADD374 pKa = 3.94PADD377 pKa = 4.1PSKK380 pKa = 11.17VILMYY385 pKa = 10.62SGEE388 pKa = 4.34SRR390 pKa = 11.84PDD392 pKa = 3.19DD393 pKa = 3.26QYY395 pKa = 12.02QLHH398 pKa = 6.44EE399 pKa = 4.53PPLRR403 pKa = 11.84GEE405 pKa = 4.39TFNTEE410 pKa = 3.91HH411 pKa = 7.62IYY413 pKa = 9.98PQSKK417 pKa = 9.96LDD419 pKa = 3.69HH420 pKa = 7.43DD421 pKa = 4.11EE422 pKa = 4.23AANDD426 pKa = 3.7LHH428 pKa = 7.69HH429 pKa = 6.62MRR431 pKa = 11.84VADD434 pKa = 3.17IDD436 pKa = 4.32VNSEE440 pKa = 3.26RR441 pKa = 11.84SYY443 pKa = 11.87YY444 pKa = 10.18PFTDD448 pKa = 3.32GTGTYY453 pKa = 10.8KK454 pKa = 10.87LVDD457 pKa = 3.63GNKK460 pKa = 8.95WFPGDD465 pKa = 3.52DD466 pKa = 3.11WRR468 pKa = 11.84GDD470 pKa = 3.45VARR473 pKa = 11.84MVMYY477 pKa = 11.02VNLAYY482 pKa = 10.23GDD484 pKa = 4.02SFDD487 pKa = 4.35EE488 pKa = 4.47VGSKK492 pKa = 10.92DD493 pKa = 6.14LFLKK497 pKa = 9.84WNRR500 pKa = 11.84EE501 pKa = 4.0DD502 pKa = 3.58PVSEE506 pKa = 5.39FEE508 pKa = 3.92IQRR511 pKa = 11.84NNVIEE516 pKa = 4.38GAQGNRR522 pKa = 11.84NPFIDD527 pKa = 3.96NPYY530 pKa = 10.19LATLIWGGEE539 pKa = 3.84PAQNRR544 pKa = 11.84WQQ546 pKa = 3.34
Molecular weight: 60.47 kDa
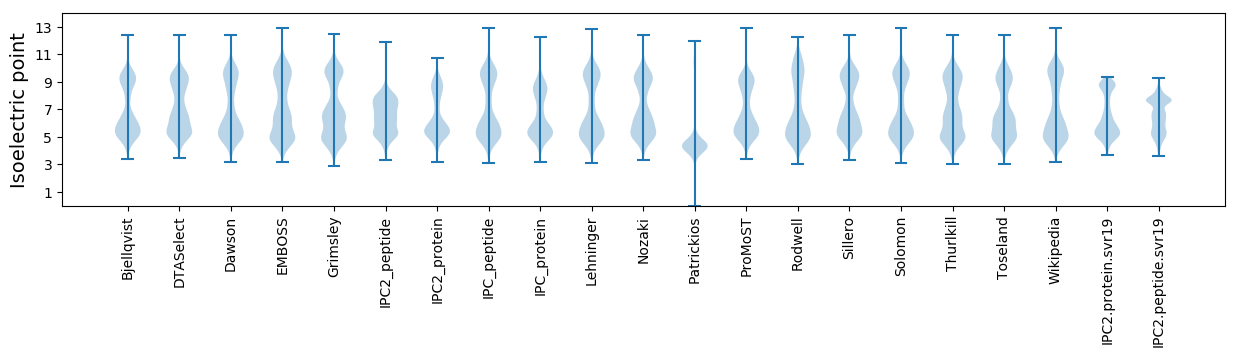
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2R3Z8W2|A0A2R3Z8W2_9FLAO Chalcone isomerase OS=Gramella fulva OX=2126553 GN=C7S20_15985 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 10.15RR10 pKa = 11.84KK11 pKa = 9.6RR12 pKa = 11.84KK13 pKa = 8.29NKK15 pKa = 9.34HH16 pKa = 4.05GFRR19 pKa = 11.84EE20 pKa = 4.21RR21 pKa = 11.84MSSANGRR28 pKa = 11.84KK29 pKa = 8.88VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.09GRR39 pKa = 11.84KK40 pKa = 7.97KK41 pKa = 10.73LSVSSEE47 pKa = 3.76NRR49 pKa = 11.84HH50 pKa = 4.47KK51 pKa = 10.5HH52 pKa = 4.49
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 10.15RR10 pKa = 11.84KK11 pKa = 9.6RR12 pKa = 11.84KK13 pKa = 8.29NKK15 pKa = 9.34HH16 pKa = 4.05GFRR19 pKa = 11.84EE20 pKa = 4.21RR21 pKa = 11.84MSSANGRR28 pKa = 11.84KK29 pKa = 8.88VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.09GRR39 pKa = 11.84KK40 pKa = 7.97KK41 pKa = 10.73LSVSSEE47 pKa = 3.76NRR49 pKa = 11.84HH50 pKa = 4.47KK51 pKa = 10.5HH52 pKa = 4.49
Molecular weight: 6.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1310285 |
25 |
12288 |
346.2 |
39.2 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.389 ± 0.05 | 0.697 ± 0.015 |
5.601 ± 0.038 | 7.354 ± 0.058 |
5.235 ± 0.036 | 6.605 ± 0.041 |
1.822 ± 0.023 | 7.593 ± 0.037 |
7.83 ± 0.077 | 9.331 ± 0.061 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.279 ± 0.028 | 5.7 ± 0.047 |
3.648 ± 0.021 | 3.383 ± 0.023 |
3.92 ± 0.048 | 6.428 ± 0.051 |
5.123 ± 0.116 | 5.898 ± 0.037 |
1.152 ± 0.02 | 4.013 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
