
Blastococcus saxobsidens
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Geodermatophilales; Geodermatophilaceae; Blastococcus
Average proteome isoelectric point is 6.1
Get precalculated fractions of proteins
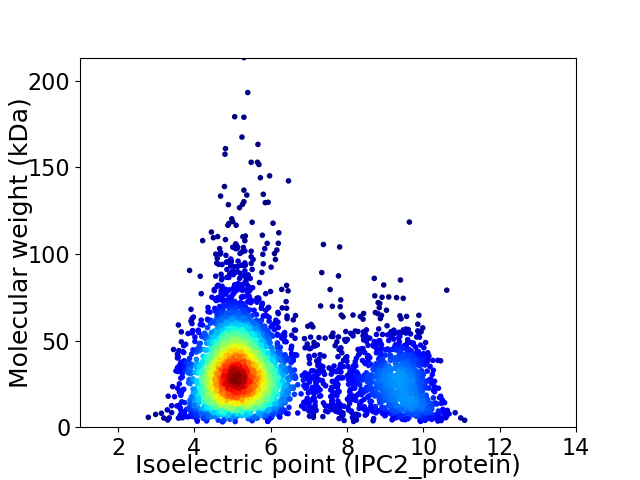
Virtual 2D-PAGE plot for 4219 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Q7Y867|A0A4Q7Y867_9ACTN Glucose/arabinose dehydrogenase OS=Blastococcus saxobsidens OX=138336 GN=BKA19_2973 PE=4 SV=1
MM1 pKa = 7.35RR2 pKa = 11.84QLLRR6 pKa = 11.84RR7 pKa = 11.84AVPALLAGVLVAGCAVDD24 pKa = 3.81VVRR27 pKa = 11.84GEE29 pKa = 4.22ATPGTSGPTDD39 pKa = 2.84IGVEE43 pKa = 4.08EE44 pKa = 4.77FPVTGASDD52 pKa = 3.61EE53 pKa = 5.32PIDD56 pKa = 3.86QFARR60 pKa = 11.84NALADD65 pKa = 4.96LEE67 pKa = 5.53SFWQTAYY74 pKa = 9.88PAFFGAEE81 pKa = 4.01YY82 pKa = 10.45QPLAGGYY89 pKa = 9.75FSVDD93 pKa = 3.32SVDD96 pKa = 4.76LDD98 pKa = 3.37ASAYY102 pKa = 8.59PEE104 pKa = 4.08TGIGCEE110 pKa = 4.25GSPTAPEE117 pKa = 4.17DD118 pKa = 3.48VAGNAFYY125 pKa = 11.11DD126 pKa = 4.06PVCDD130 pKa = 4.21VIAYY134 pKa = 8.88DD135 pKa = 3.53RR136 pKa = 11.84TLLQEE141 pKa = 4.22LADD144 pKa = 4.3DD145 pKa = 4.16YY146 pKa = 11.83GRR148 pKa = 11.84FLAPVVMAHH157 pKa = 6.21EE158 pKa = 5.55FGHH161 pKa = 6.59AMQGRR166 pKa = 11.84FGFAASGRR174 pKa = 11.84SILDD178 pKa = 3.22EE179 pKa = 4.19TQADD183 pKa = 4.22CLAGAWTRR191 pKa = 11.84WVADD195 pKa = 3.59GQARR199 pKa = 11.84YY200 pKa = 8.0VTLRR204 pKa = 11.84EE205 pKa = 4.13PEE207 pKa = 4.12LDD209 pKa = 3.52DD210 pKa = 3.65VVRR213 pKa = 11.84GFLLLRR219 pKa = 11.84DD220 pKa = 4.48DD221 pKa = 4.65VGSDD225 pKa = 3.54PDD227 pKa = 3.46DD228 pKa = 4.2TEE230 pKa = 4.14AHH232 pKa = 6.24GSFFDD237 pKa = 3.69RR238 pKa = 11.84VSAFSDD244 pKa = 3.73GFDD247 pKa = 3.64GGLAVCRR254 pKa = 11.84DD255 pKa = 3.62EE256 pKa = 5.35FGEE259 pKa = 4.57DD260 pKa = 3.14RR261 pKa = 11.84LFTAAAFTPTDD272 pKa = 3.78EE273 pKa = 4.99LSQGNAPFADD283 pKa = 3.12IVDD286 pKa = 4.3WVATTLLAFWAEE298 pKa = 4.12AFPQTFGGEE307 pKa = 4.19FEE309 pKa = 4.42PPALEE314 pKa = 4.33GFGGGAPDD322 pKa = 4.94CDD324 pKa = 3.71GLDD327 pKa = 3.56GRR329 pKa = 11.84RR330 pKa = 11.84LGYY333 pKa = 10.2CADD336 pKa = 3.71EE337 pKa = 4.04ATVYY341 pKa = 10.79VDD343 pKa = 3.48EE344 pKa = 5.12TEE346 pKa = 4.43LAVPAYY352 pKa = 10.64DD353 pKa = 3.6EE354 pKa = 4.67VGDD357 pKa = 4.11FALTTALSLPYY368 pKa = 10.34ALAVRR373 pKa = 11.84DD374 pKa = 3.78QAGLSVDD381 pKa = 4.46DD382 pKa = 4.59GAATRR387 pKa = 11.84SAVCLTGWYY396 pKa = 7.3EE397 pKa = 4.1AQWYY401 pKa = 7.74TDD403 pKa = 3.15AFADD407 pKa = 4.58LVPAQISPGDD417 pKa = 3.44IDD419 pKa = 3.79EE420 pKa = 4.94AVQFLLQYY428 pKa = 10.92GVDD431 pKa = 3.89DD432 pKa = 3.82QVLPDD437 pKa = 3.4VDD439 pKa = 3.81ASGFEE444 pKa = 4.06LVGEE448 pKa = 4.23FRR450 pKa = 11.84SGFLYY455 pKa = 10.51GAEE458 pKa = 4.29GCGLGG463 pKa = 4.38
MM1 pKa = 7.35RR2 pKa = 11.84QLLRR6 pKa = 11.84RR7 pKa = 11.84AVPALLAGVLVAGCAVDD24 pKa = 3.81VVRR27 pKa = 11.84GEE29 pKa = 4.22ATPGTSGPTDD39 pKa = 2.84IGVEE43 pKa = 4.08EE44 pKa = 4.77FPVTGASDD52 pKa = 3.61EE53 pKa = 5.32PIDD56 pKa = 3.86QFARR60 pKa = 11.84NALADD65 pKa = 4.96LEE67 pKa = 5.53SFWQTAYY74 pKa = 9.88PAFFGAEE81 pKa = 4.01YY82 pKa = 10.45QPLAGGYY89 pKa = 9.75FSVDD93 pKa = 3.32SVDD96 pKa = 4.76LDD98 pKa = 3.37ASAYY102 pKa = 8.59PEE104 pKa = 4.08TGIGCEE110 pKa = 4.25GSPTAPEE117 pKa = 4.17DD118 pKa = 3.48VAGNAFYY125 pKa = 11.11DD126 pKa = 4.06PVCDD130 pKa = 4.21VIAYY134 pKa = 8.88DD135 pKa = 3.53RR136 pKa = 11.84TLLQEE141 pKa = 4.22LADD144 pKa = 4.3DD145 pKa = 4.16YY146 pKa = 11.83GRR148 pKa = 11.84FLAPVVMAHH157 pKa = 6.21EE158 pKa = 5.55FGHH161 pKa = 6.59AMQGRR166 pKa = 11.84FGFAASGRR174 pKa = 11.84SILDD178 pKa = 3.22EE179 pKa = 4.19TQADD183 pKa = 4.22CLAGAWTRR191 pKa = 11.84WVADD195 pKa = 3.59GQARR199 pKa = 11.84YY200 pKa = 8.0VTLRR204 pKa = 11.84EE205 pKa = 4.13PEE207 pKa = 4.12LDD209 pKa = 3.52DD210 pKa = 3.65VVRR213 pKa = 11.84GFLLLRR219 pKa = 11.84DD220 pKa = 4.48DD221 pKa = 4.65VGSDD225 pKa = 3.54PDD227 pKa = 3.46DD228 pKa = 4.2TEE230 pKa = 4.14AHH232 pKa = 6.24GSFFDD237 pKa = 3.69RR238 pKa = 11.84VSAFSDD244 pKa = 3.73GFDD247 pKa = 3.64GGLAVCRR254 pKa = 11.84DD255 pKa = 3.62EE256 pKa = 5.35FGEE259 pKa = 4.57DD260 pKa = 3.14RR261 pKa = 11.84LFTAAAFTPTDD272 pKa = 3.78EE273 pKa = 4.99LSQGNAPFADD283 pKa = 3.12IVDD286 pKa = 4.3WVATTLLAFWAEE298 pKa = 4.12AFPQTFGGEE307 pKa = 4.19FEE309 pKa = 4.42PPALEE314 pKa = 4.33GFGGGAPDD322 pKa = 4.94CDD324 pKa = 3.71GLDD327 pKa = 3.56GRR329 pKa = 11.84RR330 pKa = 11.84LGYY333 pKa = 10.2CADD336 pKa = 3.71EE337 pKa = 4.04ATVYY341 pKa = 10.79VDD343 pKa = 3.48EE344 pKa = 5.12TEE346 pKa = 4.43LAVPAYY352 pKa = 10.64DD353 pKa = 3.6EE354 pKa = 4.67VGDD357 pKa = 4.11FALTTALSLPYY368 pKa = 10.34ALAVRR373 pKa = 11.84DD374 pKa = 3.78QAGLSVDD381 pKa = 4.46DD382 pKa = 4.59GAATRR387 pKa = 11.84SAVCLTGWYY396 pKa = 7.3EE397 pKa = 4.1AQWYY401 pKa = 7.74TDD403 pKa = 3.15AFADD407 pKa = 4.58LVPAQISPGDD417 pKa = 3.44IDD419 pKa = 3.79EE420 pKa = 4.94AVQFLLQYY428 pKa = 10.92GVDD431 pKa = 3.89DD432 pKa = 3.82QVLPDD437 pKa = 3.4VDD439 pKa = 3.81ASGFEE444 pKa = 4.06LVGEE448 pKa = 4.23FRR450 pKa = 11.84SGFLYY455 pKa = 10.51GAEE458 pKa = 4.29GCGLGG463 pKa = 4.38
Molecular weight: 49.3 kDa
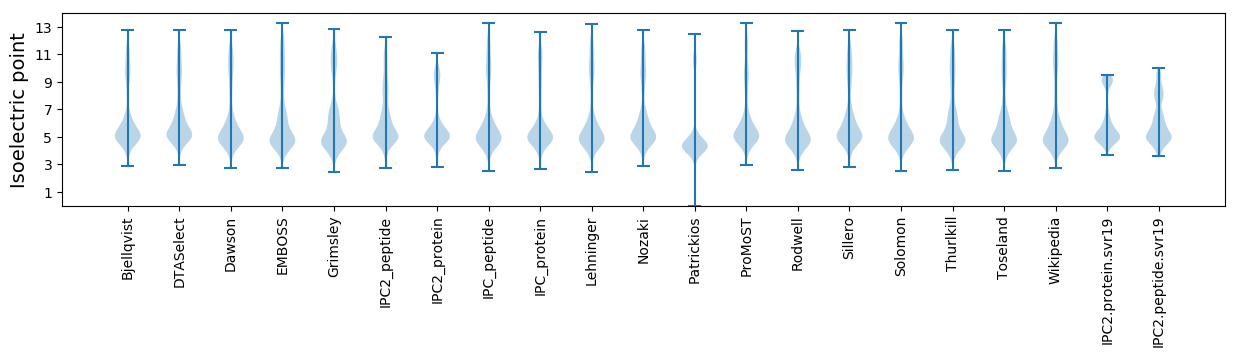
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Q7YAU2|A0A4Q7YAU2_9ACTN Uncharacterized protein DUF4397 OS=Blastococcus saxobsidens OX=138336 GN=BKA19_3693 PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.99KK16 pKa = 9.77HH17 pKa = 5.81RR18 pKa = 11.84KK19 pKa = 8.51LLKK22 pKa = 8.15KK23 pKa = 9.25TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.81KK32 pKa = 9.63
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.99KK16 pKa = 9.77HH17 pKa = 5.81RR18 pKa = 11.84KK19 pKa = 8.51LLKK22 pKa = 8.15KK23 pKa = 9.25TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.81KK32 pKa = 9.63
Molecular weight: 4.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1345434 |
30 |
1989 |
318.9 |
33.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.226 ± 0.063 | 0.677 ± 0.011 |
6.18 ± 0.031 | 5.674 ± 0.032 |
2.613 ± 0.022 | 9.71 ± 0.031 |
1.979 ± 0.019 | 2.91 ± 0.025 |
1.425 ± 0.024 | 10.716 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.684 ± 0.014 | 1.451 ± 0.016 |
6.176 ± 0.035 | 2.728 ± 0.021 |
8.091 ± 0.036 | 4.913 ± 0.023 |
5.921 ± 0.026 | 9.703 ± 0.04 |
1.465 ± 0.018 | 1.758 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
