
Halioglobus sediminis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Cellvibrionales; Halieaceae; Halioglobus
Average proteome isoelectric point is 5.88
Get precalculated fractions of proteins
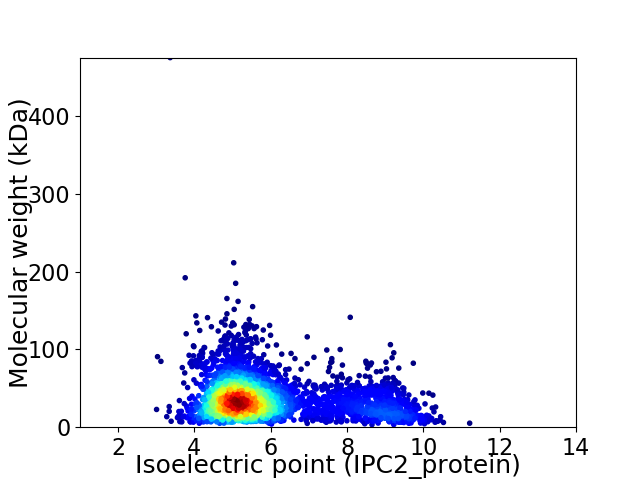
Virtual 2D-PAGE plot for 3770 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3L7DX27|A0A3L7DX27_9GAMM Dihydroxy-acid dehydratase OS=Halioglobus sediminis OX=2283635 GN=ilvD PE=3 SV=1
MM1 pKa = 7.43AVALTLLLAIAACGGGGGGGGGFKK25 pKa = 10.82GDD27 pKa = 3.72GGSGGDD33 pKa = 4.36ADD35 pKa = 4.01NDD37 pKa = 4.15TYY39 pKa = 10.99FIQLALRR46 pKa = 11.84DD47 pKa = 4.21SEE49 pKa = 4.97GEE51 pKa = 3.76PSSIVTSNSPGTLQVLVTEE70 pKa = 5.14KK71 pKa = 10.49NAKK74 pKa = 9.31GDD76 pKa = 3.86PVPDD80 pKa = 4.08VIVTASTDD88 pKa = 3.36SGLLDD93 pKa = 4.01PASGNALTNAEE104 pKa = 4.17GAVTFTVRR112 pKa = 11.84AGQDD116 pKa = 2.77RR117 pKa = 11.84GAGTIVVSVEE127 pKa = 3.91DD128 pKa = 3.62PAGVVIEE135 pKa = 4.49EE136 pKa = 4.49SVNFQIGIGDD146 pKa = 3.79LRR148 pKa = 11.84LGHH151 pKa = 6.47LQGSSFFDD159 pKa = 3.49AEE161 pKa = 4.06IGITPEE167 pKa = 3.83GPIAARR173 pKa = 11.84GEE175 pKa = 4.68AILSLAIVDD184 pKa = 4.2PKK186 pKa = 10.97GLPIGTAEE194 pKa = 4.15SIKK197 pKa = 10.68INSLCLDD204 pKa = 3.63IGDD207 pKa = 4.83ASLSPEE213 pKa = 3.9NPIPVVDD220 pKa = 4.02GRR222 pKa = 11.84VEE224 pKa = 4.04VTYY227 pKa = 9.25TAAGCQGMDD236 pKa = 3.27EE237 pKa = 4.44LTAEE241 pKa = 4.47VIGVGAQAFGTIEE254 pKa = 3.91ISDD257 pKa = 4.28TIANGLTFVSAEE269 pKa = 3.88PNLIVLRR276 pKa = 11.84GTGGGPTRR284 pKa = 11.84QEE286 pKa = 3.7KK287 pKa = 11.01SEE289 pKa = 4.1VTFQAVDD296 pKa = 3.42ANAQPLEE303 pKa = 4.22GVDD306 pKa = 3.65VVFSLTTDD314 pKa = 3.01VGGLSFSPQSATTGADD330 pKa = 3.32GTASTVVSSGDD341 pKa = 3.16VATVVRR347 pKa = 11.84VVATAEE353 pKa = 4.0ADD355 pKa = 3.66DD356 pKa = 4.78LGEE359 pKa = 4.07VSAVSDD365 pKa = 3.83VLTVSTGLPDD375 pKa = 3.37QNSISLSVDD384 pKa = 2.87GNFVVEE390 pKa = 4.51EE391 pKa = 4.41GMTKK395 pKa = 10.58DD396 pKa = 3.08GVTRR400 pKa = 11.84QLTVRR405 pKa = 11.84MADD408 pKa = 3.12KK409 pKa = 10.75FNNPVPDD416 pKa = 3.34GTAAVFTTEE425 pKa = 3.69YY426 pKa = 11.08GAIQSSCEE434 pKa = 4.12TVGGQCSVTWNSQAPRR450 pKa = 11.84VPTLNEE456 pKa = 3.77NQDD459 pKa = 3.76LVVTIFDD466 pKa = 4.45PGYY469 pKa = 10.57DD470 pKa = 3.81CPSHH474 pKa = 6.5NGSSGPCPDD483 pKa = 4.36DD484 pKa = 4.67LGMIRR489 pKa = 11.84GGRR492 pKa = 11.84STILVTAIGEE502 pKa = 4.1EE503 pKa = 4.24SFIDD507 pKa = 3.76SNANGIYY514 pKa = 10.28DD515 pKa = 3.56EE516 pKa = 4.83GEE518 pKa = 3.76RR519 pKa = 11.84FANQSEE525 pKa = 4.24AFLDD529 pKa = 3.56INEE532 pKa = 4.4NGFYY536 pKa = 10.6DD537 pKa = 4.82QSTTACNNNPDD548 pKa = 4.03TLTCKK553 pKa = 10.4AGSEE557 pKa = 4.43EE558 pKa = 4.27IFTDD562 pKa = 4.83FNSNGVFDD570 pKa = 5.81ANGDD574 pKa = 3.97DD575 pKa = 4.33EE576 pKa = 6.05LNGYY580 pKa = 8.71PDD582 pKa = 3.91EE583 pKa = 4.83GVQALYY589 pKa = 11.03NGLLCPKK596 pKa = 9.49EE597 pKa = 3.95GDD599 pKa = 3.3GVYY602 pKa = 10.44CSRR605 pKa = 11.84EE606 pKa = 3.89LLNVNDD612 pKa = 4.26SLVLILSTDD621 pKa = 3.99PNWDD625 pKa = 2.89IALYY629 pKa = 10.53RR630 pKa = 11.84GVTPANSTTYY640 pKa = 10.98NGGTYY645 pKa = 7.97TAYY648 pKa = 9.91ISDD651 pKa = 4.55YY652 pKa = 10.58YY653 pKa = 10.8NSKK656 pKa = 8.5PTGGSTVSIEE666 pKa = 3.85ASGDD670 pKa = 3.61CEE672 pKa = 4.05ITGKK676 pKa = 10.69EE677 pKa = 4.08SFEE680 pKa = 4.42VPNTTAPGAFAVSFSQGGVGEE701 pKa = 4.25EE702 pKa = 3.97AGEE705 pKa = 4.3VTVTLSPNGGGPDD718 pKa = 3.78YY719 pKa = 11.23SEE721 pKa = 4.9SWPCTPEE728 pKa = 4.04PEE730 pKa = 4.26PVEE733 pKa = 4.89PEE735 pKa = 3.87PCDD738 pKa = 3.75PNVEE742 pKa = 4.16LCAGGG747 pKa = 3.36
MM1 pKa = 7.43AVALTLLLAIAACGGGGGGGGGFKK25 pKa = 10.82GDD27 pKa = 3.72GGSGGDD33 pKa = 4.36ADD35 pKa = 4.01NDD37 pKa = 4.15TYY39 pKa = 10.99FIQLALRR46 pKa = 11.84DD47 pKa = 4.21SEE49 pKa = 4.97GEE51 pKa = 3.76PSSIVTSNSPGTLQVLVTEE70 pKa = 5.14KK71 pKa = 10.49NAKK74 pKa = 9.31GDD76 pKa = 3.86PVPDD80 pKa = 4.08VIVTASTDD88 pKa = 3.36SGLLDD93 pKa = 4.01PASGNALTNAEE104 pKa = 4.17GAVTFTVRR112 pKa = 11.84AGQDD116 pKa = 2.77RR117 pKa = 11.84GAGTIVVSVEE127 pKa = 3.91DD128 pKa = 3.62PAGVVIEE135 pKa = 4.49EE136 pKa = 4.49SVNFQIGIGDD146 pKa = 3.79LRR148 pKa = 11.84LGHH151 pKa = 6.47LQGSSFFDD159 pKa = 3.49AEE161 pKa = 4.06IGITPEE167 pKa = 3.83GPIAARR173 pKa = 11.84GEE175 pKa = 4.68AILSLAIVDD184 pKa = 4.2PKK186 pKa = 10.97GLPIGTAEE194 pKa = 4.15SIKK197 pKa = 10.68INSLCLDD204 pKa = 3.63IGDD207 pKa = 4.83ASLSPEE213 pKa = 3.9NPIPVVDD220 pKa = 4.02GRR222 pKa = 11.84VEE224 pKa = 4.04VTYY227 pKa = 9.25TAAGCQGMDD236 pKa = 3.27EE237 pKa = 4.44LTAEE241 pKa = 4.47VIGVGAQAFGTIEE254 pKa = 3.91ISDD257 pKa = 4.28TIANGLTFVSAEE269 pKa = 3.88PNLIVLRR276 pKa = 11.84GTGGGPTRR284 pKa = 11.84QEE286 pKa = 3.7KK287 pKa = 11.01SEE289 pKa = 4.1VTFQAVDD296 pKa = 3.42ANAQPLEE303 pKa = 4.22GVDD306 pKa = 3.65VVFSLTTDD314 pKa = 3.01VGGLSFSPQSATTGADD330 pKa = 3.32GTASTVVSSGDD341 pKa = 3.16VATVVRR347 pKa = 11.84VVATAEE353 pKa = 4.0ADD355 pKa = 3.66DD356 pKa = 4.78LGEE359 pKa = 4.07VSAVSDD365 pKa = 3.83VLTVSTGLPDD375 pKa = 3.37QNSISLSVDD384 pKa = 2.87GNFVVEE390 pKa = 4.51EE391 pKa = 4.41GMTKK395 pKa = 10.58DD396 pKa = 3.08GVTRR400 pKa = 11.84QLTVRR405 pKa = 11.84MADD408 pKa = 3.12KK409 pKa = 10.75FNNPVPDD416 pKa = 3.34GTAAVFTTEE425 pKa = 3.69YY426 pKa = 11.08GAIQSSCEE434 pKa = 4.12TVGGQCSVTWNSQAPRR450 pKa = 11.84VPTLNEE456 pKa = 3.77NQDD459 pKa = 3.76LVVTIFDD466 pKa = 4.45PGYY469 pKa = 10.57DD470 pKa = 3.81CPSHH474 pKa = 6.5NGSSGPCPDD483 pKa = 4.36DD484 pKa = 4.67LGMIRR489 pKa = 11.84GGRR492 pKa = 11.84STILVTAIGEE502 pKa = 4.1EE503 pKa = 4.24SFIDD507 pKa = 3.76SNANGIYY514 pKa = 10.28DD515 pKa = 3.56EE516 pKa = 4.83GEE518 pKa = 3.76RR519 pKa = 11.84FANQSEE525 pKa = 4.24AFLDD529 pKa = 3.56INEE532 pKa = 4.4NGFYY536 pKa = 10.6DD537 pKa = 4.82QSTTACNNNPDD548 pKa = 4.03TLTCKK553 pKa = 10.4AGSEE557 pKa = 4.43EE558 pKa = 4.27IFTDD562 pKa = 4.83FNSNGVFDD570 pKa = 5.81ANGDD574 pKa = 3.97DD575 pKa = 4.33EE576 pKa = 6.05LNGYY580 pKa = 8.71PDD582 pKa = 3.91EE583 pKa = 4.83GVQALYY589 pKa = 11.03NGLLCPKK596 pKa = 9.49EE597 pKa = 3.95GDD599 pKa = 3.3GVYY602 pKa = 10.44CSRR605 pKa = 11.84EE606 pKa = 3.89LLNVNDD612 pKa = 4.26SLVLILSTDD621 pKa = 3.99PNWDD625 pKa = 2.89IALYY629 pKa = 10.53RR630 pKa = 11.84GVTPANSTTYY640 pKa = 10.98NGGTYY645 pKa = 7.97TAYY648 pKa = 9.91ISDD651 pKa = 4.55YY652 pKa = 10.58YY653 pKa = 10.8NSKK656 pKa = 8.5PTGGSTVSIEE666 pKa = 3.85ASGDD670 pKa = 3.61CEE672 pKa = 4.05ITGKK676 pKa = 10.69EE677 pKa = 4.08SFEE680 pKa = 4.42VPNTTAPGAFAVSFSQGGVGEE701 pKa = 4.25EE702 pKa = 3.97AGEE705 pKa = 4.3VTVTLSPNGGGPDD718 pKa = 3.78YY719 pKa = 11.23SEE721 pKa = 4.9SWPCTPEE728 pKa = 4.04PEE730 pKa = 4.26PVEE733 pKa = 4.89PEE735 pKa = 3.87PCDD738 pKa = 3.75PNVEE742 pKa = 4.16LCAGGG747 pKa = 3.36
Molecular weight: 76.58 kDa
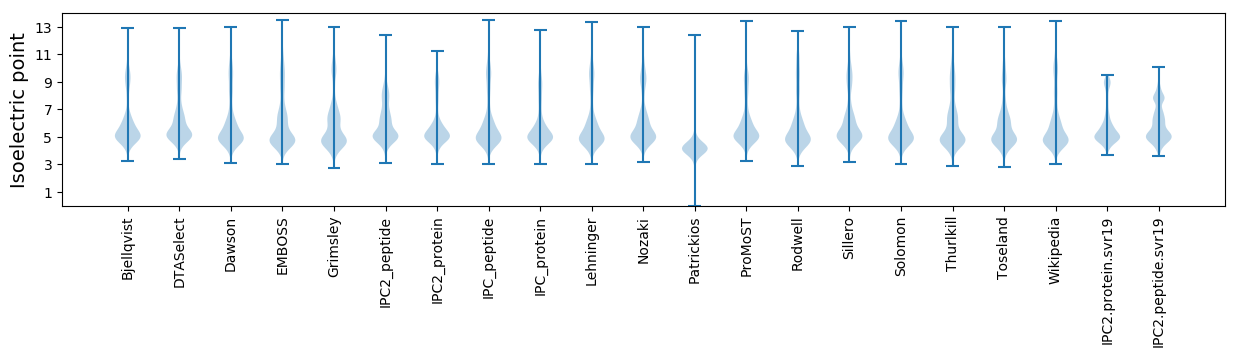
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3L7DXU2|A0A3L7DXU2_9GAMM GTP 3' 8-cyclase OS=Halioglobus sediminis OX=2283635 GN=moaA PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.49RR12 pKa = 11.84KK13 pKa = 7.87RR14 pKa = 11.84THH16 pKa = 5.99GFRR19 pKa = 11.84SRR21 pKa = 11.84MATKK25 pKa = 10.47NGRR28 pKa = 11.84AVLNRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.74GRR39 pKa = 11.84KK40 pKa = 8.67VLSAA44 pKa = 4.05
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.49RR12 pKa = 11.84KK13 pKa = 7.87RR14 pKa = 11.84THH16 pKa = 5.99GFRR19 pKa = 11.84SRR21 pKa = 11.84MATKK25 pKa = 10.47NGRR28 pKa = 11.84AVLNRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.74GRR39 pKa = 11.84KK40 pKa = 8.67VLSAA44 pKa = 4.05
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1298764 |
29 |
4582 |
344.5 |
37.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.081 ± 0.052 | 1.06 ± 0.015 |
5.944 ± 0.038 | 6.322 ± 0.036 |
3.569 ± 0.025 | 8.156 ± 0.035 |
2.149 ± 0.017 | 4.904 ± 0.028 |
2.966 ± 0.03 | 10.8 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.437 ± 0.02 | 3.017 ± 0.023 |
4.776 ± 0.026 | 4.144 ± 0.024 |
6.691 ± 0.036 | 5.897 ± 0.024 |
4.786 ± 0.027 | 7.078 ± 0.032 |
1.4 ± 0.019 | 2.822 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
