
Penaeus vannamei (Whiteleg shrimp) (Litopenaeus vannamei)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Ecdysozoa; Panarthropoda; Arthropoda; Mandibulata; Pancrustacea; Crustacea; Multicrustacea; Malacostraca; Eumalacostraca; Eucarida; Decapoda; Dendrobranchiata;
Average proteome isoelectric point is 7.22
Get precalculated fractions of proteins
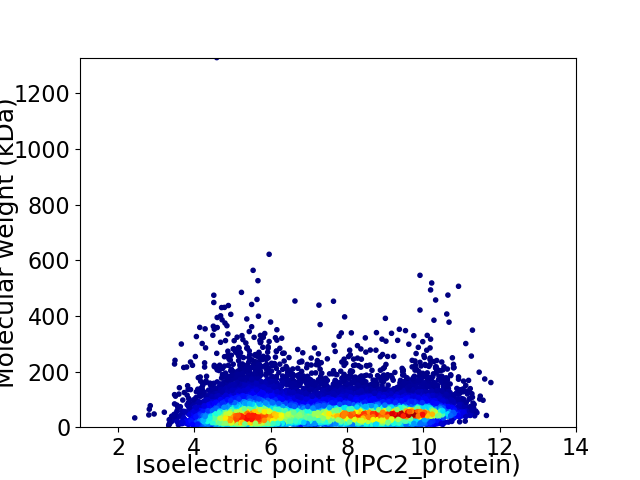
Virtual 2D-PAGE plot for 25399 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A423TD78|A0A423TD78_PENVA Uncharacterized protein OS=Penaeus vannamei OX=6689 GN=C7M84_007163 PE=3 SV=1
MM1 pKa = 7.28FLSSHH6 pKa = 7.06LCPLSYY12 pKa = 10.96SSLSLFLDD20 pKa = 3.96SLSSPLSCLALPWLEE35 pKa = 3.82ITYY38 pKa = 7.52EE39 pKa = 4.08TTDD42 pKa = 3.08GKK44 pKa = 11.2KK45 pKa = 8.81FTVQGKK51 pKa = 9.77EE52 pKa = 3.69GDD54 pKa = 3.68NLLDD58 pKa = 3.31IVINNDD64 pKa = 2.46VDD66 pKa = 4.62LDD68 pKa = 4.04GFGACEE74 pKa = 3.88GTLACSTCHH83 pKa = 7.38LIFKK87 pKa = 10.57KK88 pKa = 10.58EE89 pKa = 3.86DD90 pKa = 3.24FDD92 pKa = 6.26RR93 pKa = 11.84IEE95 pKa = 4.32EE96 pKa = 4.27PSTDD100 pKa = 3.85EE101 pKa = 4.17EE102 pKa = 6.21LDD104 pKa = 3.8MLDD107 pKa = 3.84LAYY110 pKa = 10.69GLTDD114 pKa = 3.18TT115 pKa = 5.38
MM1 pKa = 7.28FLSSHH6 pKa = 7.06LCPLSYY12 pKa = 10.96SSLSLFLDD20 pKa = 3.96SLSSPLSCLALPWLEE35 pKa = 3.82ITYY38 pKa = 7.52EE39 pKa = 4.08TTDD42 pKa = 3.08GKK44 pKa = 11.2KK45 pKa = 8.81FTVQGKK51 pKa = 9.77EE52 pKa = 3.69GDD54 pKa = 3.68NLLDD58 pKa = 3.31IVINNDD64 pKa = 2.46VDD66 pKa = 4.62LDD68 pKa = 4.04GFGACEE74 pKa = 3.88GTLACSTCHH83 pKa = 7.38LIFKK87 pKa = 10.57KK88 pKa = 10.58EE89 pKa = 3.86DD90 pKa = 3.24FDD92 pKa = 6.26RR93 pKa = 11.84IEE95 pKa = 4.32EE96 pKa = 4.27PSTDD100 pKa = 3.85EE101 pKa = 4.17EE102 pKa = 6.21LDD104 pKa = 3.8MLDD107 pKa = 3.84LAYY110 pKa = 10.69GLTDD114 pKa = 3.18TT115 pKa = 5.38
Molecular weight: 12.71 kDa
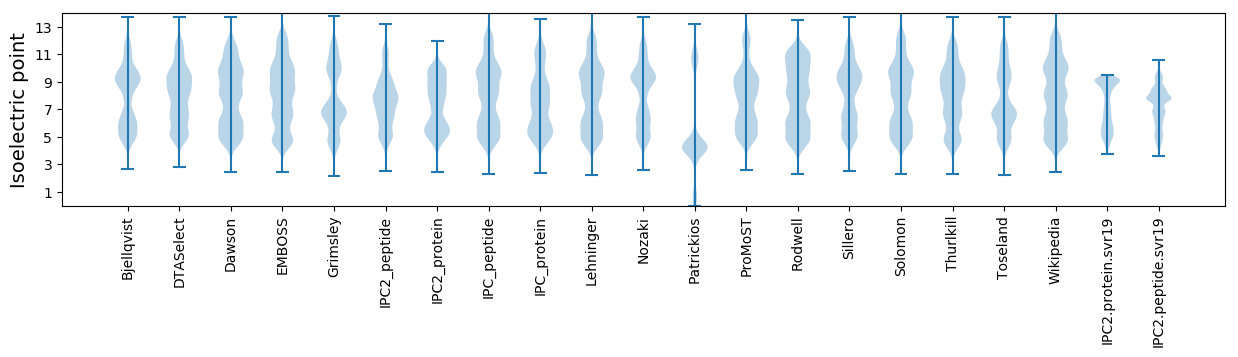
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A423U557|A0A423U557_PENVA Crustacyanin subunit A OS=Penaeus vannamei OX=6689 GN=C7M84_023007 PE=3 SV=1
MM1 pKa = 7.62VDD3 pKa = 3.32EE4 pKa = 4.64VKK6 pKa = 10.39SARR9 pKa = 11.84RR10 pKa = 11.84KK11 pKa = 9.71VLSLHH16 pKa = 6.79LSLHH20 pKa = 6.47LSASPLSPLASPLASPPRR38 pKa = 11.84PLRR41 pKa = 11.84FTLATSPSLNLSLHH55 pKa = 6.39LSLFTSRR62 pKa = 11.84LHH64 pKa = 6.66LSLPSRR70 pKa = 11.84SPLRR74 pKa = 11.84FTSRR78 pKa = 11.84SLASTLSALDD88 pKa = 3.44TSRR91 pKa = 11.84LTFLALSPLASSLQTSRR108 pKa = 11.84FHH110 pKa = 6.65LRR112 pKa = 11.84SSPSPLASPLVHH124 pKa = 6.9LSLHH128 pKa = 6.42LSFTSRR134 pKa = 11.84FTSRR138 pKa = 11.84FTSRR142 pKa = 11.84FTSRR146 pKa = 11.84TSRR149 pKa = 11.84FNLSRR154 pKa = 11.84SALFASPLAQPLASSFTSRR173 pKa = 11.84QLRR176 pKa = 11.84SPLASPLSFTSRR188 pKa = 11.84LLSLHH193 pKa = 6.69LSLHH197 pKa = 6.16LVASHH202 pKa = 7.19LSLTSRR208 pKa = 11.84FTSRR212 pKa = 11.84PPSRR216 pKa = 11.84FISLHH221 pKa = 5.19PSRR224 pKa = 11.84FNLSSPLASPLVSPLASPLVHH245 pKa = 6.82LLASRR250 pKa = 11.84SRR252 pKa = 11.84FTSRR256 pKa = 11.84FTSPLVSTSSLHH268 pKa = 6.84LSLHH272 pKa = 6.46LSLSRR277 pKa = 11.84PLASPHH283 pKa = 6.23LRR285 pKa = 11.84FTFVRR290 pKa = 11.84LSLSPLASPLLQHH303 pKa = 6.66SRR305 pKa = 11.84FTSRR309 pKa = 11.84FTSRR313 pKa = 11.84FISSRR318 pKa = 11.84SSLHH322 pKa = 6.42LSLHH326 pKa = 6.39LSLHH330 pKa = 5.96LASPLASPLASPLASPLASPLASPLASPLASPLASPLASPLASPLALAFTSRR382 pKa = 11.84ASPPLTSRR390 pKa = 11.84FTSRR394 pKa = 11.84FTSLALTSRR403 pKa = 11.84FTSRR407 pKa = 11.84FTSRR411 pKa = 11.84SPLSSPLSPLASPLASPSRR430 pKa = 11.84FTSRR434 pKa = 11.84FTSLHH439 pKa = 6.16LSLTSRR445 pKa = 11.84FTSRR449 pKa = 11.84FTSRR453 pKa = 11.84FTLAFTSRR461 pKa = 11.84SSPLFLASPLAFSLSLRR478 pKa = 11.84LSLHH482 pKa = 6.49LSLHH486 pKa = 6.37LAPLASTSASRR497 pKa = 11.84LTLTSRR503 pKa = 11.84FTLASPSSLLASPLASPLVSPLASLSLHH531 pKa = 5.6FTLSLHH537 pKa = 6.91LSLHH541 pKa = 6.03LCFTSLHH548 pKa = 6.2LSLHH552 pKa = 6.3LSLHH556 pKa = 6.37LSLHH560 pKa = 6.67LSPHH564 pKa = 6.25SPLASPLARR573 pKa = 11.84SRR575 pKa = 11.84FASRR579 pKa = 11.84SPLRR583 pKa = 11.84SSRR586 pKa = 11.84FNLSPQLSFTSRR598 pKa = 11.84FTSRR602 pKa = 11.84SPSSPLASPLARR614 pKa = 11.84LSLLSLTSRR623 pKa = 11.84FHH625 pKa = 7.51LSLLTSRR632 pKa = 11.84FTLASPLLHH641 pKa = 7.18LSLTSLAFTSRR652 pKa = 11.84FHH654 pKa = 6.91PLGFTSRR661 pKa = 11.84FTLSLSPLASSSSLRR676 pKa = 11.84SPSPLVSPLVSPLASPSLSPLVHH699 pKa = 6.83LSLTSRR705 pKa = 11.84FPSLASTSRR714 pKa = 11.84FTSRR718 pKa = 11.84FTSPASPLVSPLASPLASPLAFSRR742 pKa = 11.84FTSRR746 pKa = 11.84FTSRR750 pKa = 11.84FLALFSSFPAGG761 pKa = 3.22
MM1 pKa = 7.62VDD3 pKa = 3.32EE4 pKa = 4.64VKK6 pKa = 10.39SARR9 pKa = 11.84RR10 pKa = 11.84KK11 pKa = 9.71VLSLHH16 pKa = 6.79LSLHH20 pKa = 6.47LSASPLSPLASPLASPPRR38 pKa = 11.84PLRR41 pKa = 11.84FTLATSPSLNLSLHH55 pKa = 6.39LSLFTSRR62 pKa = 11.84LHH64 pKa = 6.66LSLPSRR70 pKa = 11.84SPLRR74 pKa = 11.84FTSRR78 pKa = 11.84SLASTLSALDD88 pKa = 3.44TSRR91 pKa = 11.84LTFLALSPLASSLQTSRR108 pKa = 11.84FHH110 pKa = 6.65LRR112 pKa = 11.84SSPSPLASPLVHH124 pKa = 6.9LSLHH128 pKa = 6.42LSFTSRR134 pKa = 11.84FTSRR138 pKa = 11.84FTSRR142 pKa = 11.84FTSRR146 pKa = 11.84TSRR149 pKa = 11.84FNLSRR154 pKa = 11.84SALFASPLAQPLASSFTSRR173 pKa = 11.84QLRR176 pKa = 11.84SPLASPLSFTSRR188 pKa = 11.84LLSLHH193 pKa = 6.69LSLHH197 pKa = 6.16LVASHH202 pKa = 7.19LSLTSRR208 pKa = 11.84FTSRR212 pKa = 11.84PPSRR216 pKa = 11.84FISLHH221 pKa = 5.19PSRR224 pKa = 11.84FNLSSPLASPLVSPLASPLVHH245 pKa = 6.82LLASRR250 pKa = 11.84SRR252 pKa = 11.84FTSRR256 pKa = 11.84FTSPLVSTSSLHH268 pKa = 6.84LSLHH272 pKa = 6.46LSLSRR277 pKa = 11.84PLASPHH283 pKa = 6.23LRR285 pKa = 11.84FTFVRR290 pKa = 11.84LSLSPLASPLLQHH303 pKa = 6.66SRR305 pKa = 11.84FTSRR309 pKa = 11.84FTSRR313 pKa = 11.84FISSRR318 pKa = 11.84SSLHH322 pKa = 6.42LSLHH326 pKa = 6.39LSLHH330 pKa = 5.96LASPLASPLASPLASPLASPLASPLASPLASPLASPLASPLASPLALAFTSRR382 pKa = 11.84ASPPLTSRR390 pKa = 11.84FTSRR394 pKa = 11.84FTSLALTSRR403 pKa = 11.84FTSRR407 pKa = 11.84FTSRR411 pKa = 11.84SPLSSPLSPLASPLASPSRR430 pKa = 11.84FTSRR434 pKa = 11.84FTSLHH439 pKa = 6.16LSLTSRR445 pKa = 11.84FTSRR449 pKa = 11.84FTSRR453 pKa = 11.84FTLAFTSRR461 pKa = 11.84SSPLFLASPLAFSLSLRR478 pKa = 11.84LSLHH482 pKa = 6.49LSLHH486 pKa = 6.37LAPLASTSASRR497 pKa = 11.84LTLTSRR503 pKa = 11.84FTLASPSSLLASPLASPLVSPLASLSLHH531 pKa = 5.6FTLSLHH537 pKa = 6.91LSLHH541 pKa = 6.03LCFTSLHH548 pKa = 6.2LSLHH552 pKa = 6.3LSLHH556 pKa = 6.37LSLHH560 pKa = 6.67LSPHH564 pKa = 6.25SPLASPLARR573 pKa = 11.84SRR575 pKa = 11.84FASRR579 pKa = 11.84SPLRR583 pKa = 11.84SSRR586 pKa = 11.84FNLSPQLSFTSRR598 pKa = 11.84FTSRR602 pKa = 11.84SPSSPLASPLARR614 pKa = 11.84LSLLSLTSRR623 pKa = 11.84FHH625 pKa = 7.51LSLLTSRR632 pKa = 11.84FTLASPLLHH641 pKa = 7.18LSLTSLAFTSRR652 pKa = 11.84FHH654 pKa = 6.91PLGFTSRR661 pKa = 11.84FTLSLSPLASSSSLRR676 pKa = 11.84SPSPLVSPLVSPLASPSLSPLVHH699 pKa = 6.83LSLTSRR705 pKa = 11.84FPSLASTSRR714 pKa = 11.84FTSRR718 pKa = 11.84FTSPASPLVSPLASPLASPLAFSRR742 pKa = 11.84FTSRR746 pKa = 11.84FTSRR750 pKa = 11.84FLALFSSFPAGG761 pKa = 3.22
Molecular weight: 81.92 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
13085305 |
52 |
11752 |
515.2 |
56.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.592 ± 0.025 | 1.915 ± 0.012 |
4.093 ± 0.02 | 5.183 ± 0.035 |
4.316 ± 0.029 | 5.657 ± 0.029 |
3.201 ± 0.021 | 3.736 ± 0.016 |
3.929 ± 0.024 | 10.781 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.685 ± 0.009 | 3.055 ± 0.016 |
10.108 ± 0.064 | 3.702 ± 0.021 |
6.188 ± 0.028 | 10.942 ± 0.046 |
6.016 ± 0.033 | 5.499 ± 0.023 |
1.039 ± 0.007 | 2.357 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
