
Talaromyces rugulosus
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina;
Average proteome isoelectric point is 6.23
Get precalculated fractions of proteins
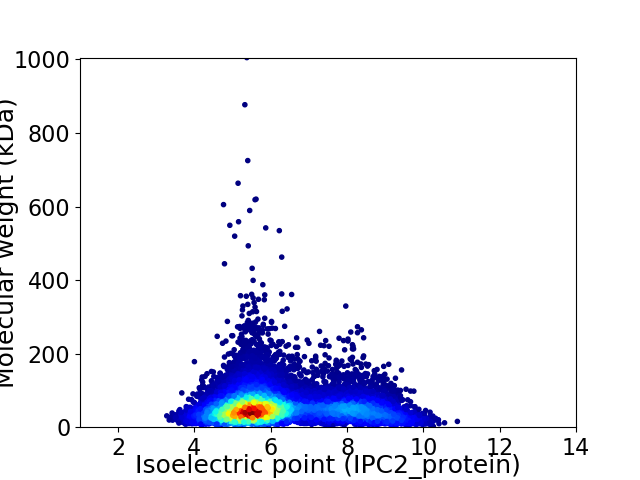
Virtual 2D-PAGE plot for 11830 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A7H8QK42|A0A7H8QK42_9EURO Uncharacterized protein OS=Talaromyces rugulosus OX=121627 GN=TRUGW13939_01351 PE=3 SV=1
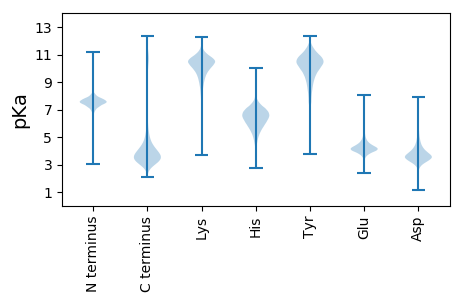
MM1 pKa = 7.83AKK3 pKa = 9.73TIFGSLVLAVVLKK16 pKa = 10.62LATANPVAQPAITSAPNLLEE36 pKa = 4.53KK37 pKa = 10.47RR38 pKa = 11.84AASCTFSGSDD48 pKa = 3.43GAASASASQKK58 pKa = 9.18SCSTIVLSAVAVPSGTTLDD77 pKa = 3.99LSNLEE82 pKa = 4.95DD83 pKa = 3.72DD84 pKa = 3.78TTVIFEE90 pKa = 5.05GEE92 pKa = 4.45TTWGYY97 pKa = 9.82EE98 pKa = 3.95EE99 pKa = 4.42WDD101 pKa = 3.96GPLLQISGTGITVQGASGASLNPDD125 pKa = 3.45GARR128 pKa = 11.84WWDD131 pKa = 3.64GEE133 pKa = 4.05GSNDD137 pKa = 4.0GKK139 pKa = 9.34TKK141 pKa = 10.47PKK143 pKa = 10.29FFYY146 pKa = 10.74AHH148 pKa = 7.64DD149 pKa = 4.2LTSSSITDD157 pKa = 3.48LQIQNTPVQAVSINGCDD174 pKa = 3.55GLTITDD180 pKa = 3.54MTIDD184 pKa = 3.5NSAGDD189 pKa = 3.81SEE191 pKa = 5.43GGHH194 pKa = 5.48NTDD197 pKa = 3.99GFDD200 pKa = 3.29IGSSSNVTIIGANVYY215 pKa = 10.16NQDD218 pKa = 3.1DD219 pKa = 4.24CVAVNSGTDD228 pKa = 3.08ITFSGGVCSGGHH240 pKa = 5.7GLSIGSVGGRR250 pKa = 11.84DD251 pKa = 4.03DD252 pKa = 3.81NTVDD256 pKa = 3.02TVTFEE261 pKa = 3.94NSEE264 pKa = 4.22VKK266 pKa = 10.78DD267 pKa = 3.68SVNGIRR273 pKa = 11.84IKK275 pKa = 10.81ASEE278 pKa = 4.49GDD280 pKa = 3.27TGTITGVTYY289 pKa = 10.81SGITLEE295 pKa = 4.72SISNRR300 pKa = 11.84ARR302 pKa = 11.84YY303 pKa = 8.76GVLIEE308 pKa = 3.96QNYY311 pKa = 10.96DD312 pKa = 3.31GGDD315 pKa = 3.66LNGGDD320 pKa = 4.18PTSGIPITDD329 pKa = 3.52LTIEE333 pKa = 4.59NISGDD338 pKa = 3.73GAVASDD344 pKa = 4.52GYY346 pKa = 11.22NIVIVCGSDD355 pKa = 3.28ACSDD359 pKa = 3.73WTWDD363 pKa = 3.69SVDD366 pKa = 3.35VTGGEE371 pKa = 4.68DD372 pKa = 4.3YY373 pKa = 11.34SDD375 pKa = 3.85CTNVPSVVSCSTT387 pKa = 3.13
MM1 pKa = 7.83AKK3 pKa = 9.73TIFGSLVLAVVLKK16 pKa = 10.62LATANPVAQPAITSAPNLLEE36 pKa = 4.53KK37 pKa = 10.47RR38 pKa = 11.84AASCTFSGSDD48 pKa = 3.43GAASASASQKK58 pKa = 9.18SCSTIVLSAVAVPSGTTLDD77 pKa = 3.99LSNLEE82 pKa = 4.95DD83 pKa = 3.72DD84 pKa = 3.78TTVIFEE90 pKa = 5.05GEE92 pKa = 4.45TTWGYY97 pKa = 9.82EE98 pKa = 3.95EE99 pKa = 4.42WDD101 pKa = 3.96GPLLQISGTGITVQGASGASLNPDD125 pKa = 3.45GARR128 pKa = 11.84WWDD131 pKa = 3.64GEE133 pKa = 4.05GSNDD137 pKa = 4.0GKK139 pKa = 9.34TKK141 pKa = 10.47PKK143 pKa = 10.29FFYY146 pKa = 10.74AHH148 pKa = 7.64DD149 pKa = 4.2LTSSSITDD157 pKa = 3.48LQIQNTPVQAVSINGCDD174 pKa = 3.55GLTITDD180 pKa = 3.54MTIDD184 pKa = 3.5NSAGDD189 pKa = 3.81SEE191 pKa = 5.43GGHH194 pKa = 5.48NTDD197 pKa = 3.99GFDD200 pKa = 3.29IGSSSNVTIIGANVYY215 pKa = 10.16NQDD218 pKa = 3.1DD219 pKa = 4.24CVAVNSGTDD228 pKa = 3.08ITFSGGVCSGGHH240 pKa = 5.7GLSIGSVGGRR250 pKa = 11.84DD251 pKa = 4.03DD252 pKa = 3.81NTVDD256 pKa = 3.02TVTFEE261 pKa = 3.94NSEE264 pKa = 4.22VKK266 pKa = 10.78DD267 pKa = 3.68SVNGIRR273 pKa = 11.84IKK275 pKa = 10.81ASEE278 pKa = 4.49GDD280 pKa = 3.27TGTITGVTYY289 pKa = 10.81SGITLEE295 pKa = 4.72SISNRR300 pKa = 11.84ARR302 pKa = 11.84YY303 pKa = 8.76GVLIEE308 pKa = 3.96QNYY311 pKa = 10.96DD312 pKa = 3.31GGDD315 pKa = 3.66LNGGDD320 pKa = 4.18PTSGIPITDD329 pKa = 3.52LTIEE333 pKa = 4.59NISGDD338 pKa = 3.73GAVASDD344 pKa = 4.52GYY346 pKa = 11.22NIVIVCGSDD355 pKa = 3.28ACSDD359 pKa = 3.73WTWDD363 pKa = 3.69SVDD366 pKa = 3.35VTGGEE371 pKa = 4.68DD372 pKa = 4.3YY373 pKa = 11.34SDD375 pKa = 3.85CTNVPSVVSCSTT387 pKa = 3.13
Molecular weight: 39.56 kDa
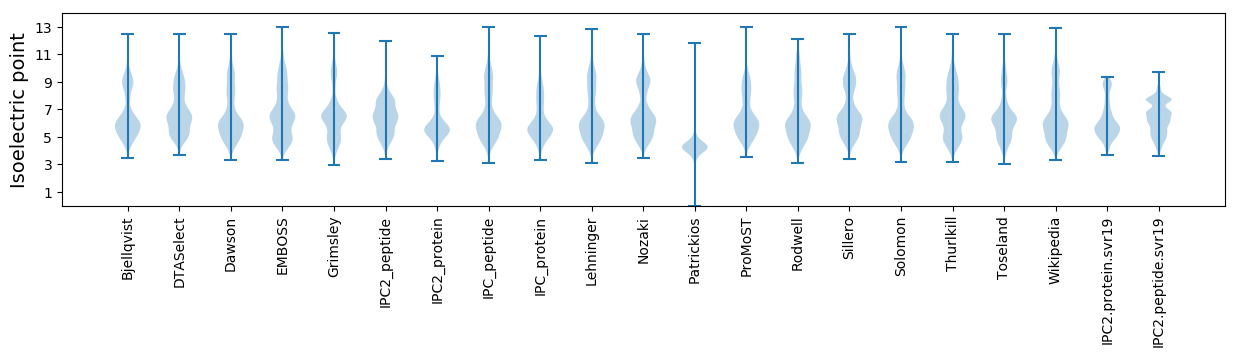
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A7H8R0R0|A0A7H8R0R0_9EURO DNL-type domain-containing protein OS=Talaromyces rugulosus OX=121627 GN=TRUGW13939_06469 PE=4 SV=1
MM1 pKa = 7.55TSHH4 pKa = 7.37SDD6 pKa = 3.46PQKK9 pKa = 10.92DD10 pKa = 3.52PNSRR14 pKa = 11.84VPLAPVLHH22 pKa = 6.24RR23 pKa = 11.84VWATSTPYY31 pKa = 9.52TASIQANIRR40 pKa = 11.84SLDD43 pKa = 3.24PRR45 pKa = 11.84RR46 pKa = 11.84PHH48 pKa = 7.02VGAHH52 pKa = 6.37HH53 pKa = 6.99SDD55 pKa = 2.76HH56 pKa = 5.75TQRR59 pKa = 11.84ARR61 pKa = 11.84YY62 pKa = 9.18IGGPSATLLLHH73 pKa = 7.22DD74 pKa = 5.55LLSCSSPACRR84 pKa = 11.84PAAQPGPRR92 pKa = 11.84AHH94 pKa = 7.44RR95 pKa = 11.84NQPGQGTRR103 pKa = 11.84SRR105 pKa = 11.84CKK107 pKa = 9.4YY108 pKa = 9.21VRR110 pKa = 11.84RR111 pKa = 11.84HH112 pKa = 4.94NALPNRR118 pKa = 11.84VCAKK122 pKa = 10.43LGALVPTGSRR132 pKa = 11.84VAATIPMFPRR142 pKa = 11.84EE143 pKa = 3.96HH144 pKa = 6.38VEE146 pKa = 3.83CVEE149 pKa = 4.07RR150 pKa = 11.84SEE152 pKa = 4.19HH153 pKa = 5.33TKK155 pKa = 10.6CLEE158 pKa = 4.02PGEE161 pKa = 4.39RR162 pKa = 11.84RR163 pKa = 11.84NADD166 pKa = 3.12SS167 pKa = 3.78
MM1 pKa = 7.55TSHH4 pKa = 7.37SDD6 pKa = 3.46PQKK9 pKa = 10.92DD10 pKa = 3.52PNSRR14 pKa = 11.84VPLAPVLHH22 pKa = 6.24RR23 pKa = 11.84VWATSTPYY31 pKa = 9.52TASIQANIRR40 pKa = 11.84SLDD43 pKa = 3.24PRR45 pKa = 11.84RR46 pKa = 11.84PHH48 pKa = 7.02VGAHH52 pKa = 6.37HH53 pKa = 6.99SDD55 pKa = 2.76HH56 pKa = 5.75TQRR59 pKa = 11.84ARR61 pKa = 11.84YY62 pKa = 9.18IGGPSATLLLHH73 pKa = 7.22DD74 pKa = 5.55LLSCSSPACRR84 pKa = 11.84PAAQPGPRR92 pKa = 11.84AHH94 pKa = 7.44RR95 pKa = 11.84NQPGQGTRR103 pKa = 11.84SRR105 pKa = 11.84CKK107 pKa = 9.4YY108 pKa = 9.21VRR110 pKa = 11.84RR111 pKa = 11.84HH112 pKa = 4.94NALPNRR118 pKa = 11.84VCAKK122 pKa = 10.43LGALVPTGSRR132 pKa = 11.84VAATIPMFPRR142 pKa = 11.84EE143 pKa = 3.96HH144 pKa = 6.38VEE146 pKa = 3.83CVEE149 pKa = 4.07RR150 pKa = 11.84SEE152 pKa = 4.19HH153 pKa = 5.33TKK155 pKa = 10.6CLEE158 pKa = 4.02PGEE161 pKa = 4.39RR162 pKa = 11.84RR163 pKa = 11.84NADD166 pKa = 3.12SS167 pKa = 3.78
Molecular weight: 18.37 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6275300 |
12 |
9054 |
530.5 |
58.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.249 ± 0.019 | 1.221 ± 0.007 |
5.745 ± 0.014 | 6.035 ± 0.022 |
3.89 ± 0.014 | 6.68 ± 0.02 |
2.415 ± 0.01 | 5.283 ± 0.013 |
4.726 ± 0.019 | 9.093 ± 0.023 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.088 ± 0.008 | 3.959 ± 0.01 |
5.715 ± 0.024 | 4.102 ± 0.012 |
5.767 ± 0.016 | 8.428 ± 0.026 |
6.041 ± 0.014 | 6.162 ± 0.015 |
1.499 ± 0.009 | 2.905 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
