
Pandoravirus inopinatum
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified dsDNA viruses; Pandoravirus
Average proteome isoelectric point is 7.39
Get precalculated fractions of proteins
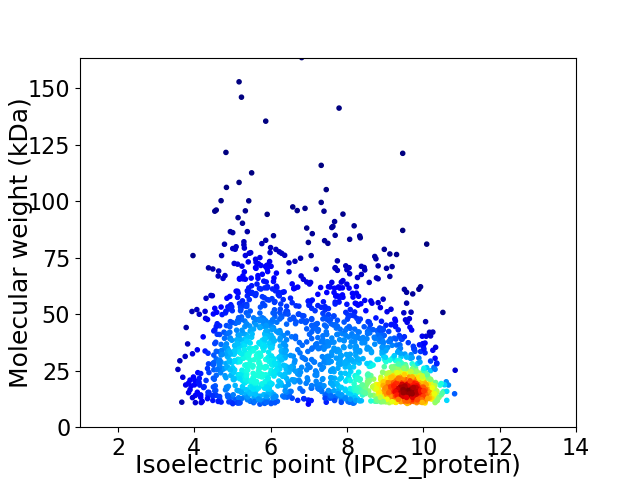
Virtual 2D-PAGE plot for 1839 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B5IYE6|A0A0B5IYE6_9VIRU Uncharacterized protein OS=Pandoravirus inopinatum OX=1605721 PE=4 SV=1
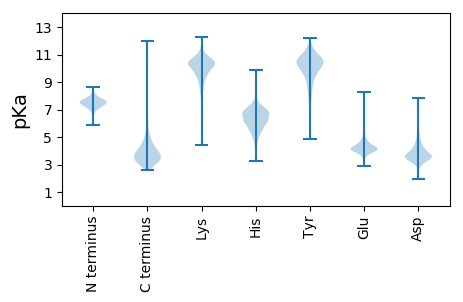
MM1 pKa = 7.02TALGKK6 pKa = 10.46RR7 pKa = 11.84ILEE10 pKa = 4.1IVSTRR15 pKa = 11.84VEE17 pKa = 4.58DD18 pKa = 4.84DD19 pKa = 4.12DD20 pKa = 4.1GTPAVPLAKK29 pKa = 9.93RR30 pKa = 11.84PRR32 pKa = 11.84CDD34 pKa = 3.34NMPEE38 pKa = 4.18PGCAGIAVGDD48 pKa = 3.55TWNGIEE54 pKa = 4.69CDD56 pKa = 3.81EE57 pKa = 4.74EE58 pKa = 4.51DD59 pKa = 4.41VEE61 pKa = 4.61ARR63 pKa = 11.84DD64 pKa = 4.1HH65 pKa = 6.03IVEE68 pKa = 4.22VVLAGGALATVLDD81 pKa = 4.23QASCHH86 pKa = 6.51HH87 pKa = 7.13IDD89 pKa = 4.07LALIQLFDD97 pKa = 4.43DD98 pKa = 4.06CTALARR104 pKa = 11.84LCRR107 pKa = 11.84AIAPSLPQALPAVLAADD124 pKa = 4.09SEE126 pKa = 4.49PDD128 pKa = 2.93KK129 pKa = 10.91TVYY132 pKa = 10.38HH133 pKa = 5.02VTIDD137 pKa = 3.76ALLCRR142 pKa = 11.84AAQQGRR148 pKa = 11.84PAVLAVLMRR157 pKa = 11.84HH158 pKa = 6.01LADD161 pKa = 4.24EE162 pKa = 4.44YY163 pKa = 11.35DD164 pKa = 3.49VARR167 pKa = 11.84ALEE170 pKa = 4.2MADD173 pKa = 4.7EE174 pKa = 4.71EE175 pKa = 4.97DD176 pKa = 4.27DD177 pKa = 3.86PAAIEE182 pKa = 4.99LIAEE186 pKa = 4.49TYY188 pKa = 10.41EE189 pKa = 4.09KK190 pKa = 10.87DD191 pKa = 3.58AGKK194 pKa = 10.34DD195 pKa = 3.27DD196 pKa = 3.79TGEE199 pKa = 4.72DD200 pKa = 3.67DD201 pKa = 4.04TGEE204 pKa = 4.77DD205 pKa = 3.68DD206 pKa = 3.75TGEE209 pKa = 4.69DD210 pKa = 3.38EE211 pKa = 4.4TGSCRR216 pKa = 11.84RR217 pKa = 11.84IAYY220 pKa = 8.84VALTCAVHH228 pKa = 6.23SSRR231 pKa = 11.84VALIHH236 pKa = 6.86RR237 pKa = 11.84LVDD240 pKa = 3.14KK241 pKa = 11.17CDD243 pKa = 3.4VEE245 pKa = 5.08DD246 pKa = 3.67VHH248 pKa = 8.1GLLDD252 pKa = 4.45DD253 pKa = 4.32CLEE256 pKa = 4.9DD257 pKa = 4.78GDD259 pKa = 4.59VFAALWQHH267 pKa = 6.65AVLCAHH273 pKa = 7.62AYY275 pKa = 8.98AASLPPCPALDD286 pKa = 3.72HH287 pKa = 6.35LTLQIANGEE296 pKa = 4.19PCADD300 pKa = 3.17MCASYY305 pKa = 10.99EE306 pKa = 4.15PADD309 pKa = 4.17SDD311 pKa = 5.13NDD313 pKa = 3.99TDD315 pKa = 6.49DD316 pKa = 5.31DD317 pKa = 4.39TDD319 pKa = 5.77DD320 pKa = 6.2DD321 pKa = 5.5DD322 pKa = 7.18GGDD325 pKa = 4.1NGNDD329 pKa = 4.38DD330 pKa = 5.53DD331 pKa = 6.73DD332 pKa = 6.51NDD334 pKa = 4.33DD335 pKa = 4.17ADD337 pKa = 4.06QGSHH341 pKa = 5.42EE342 pKa = 4.23QAQQ345 pKa = 3.42
MM1 pKa = 7.02TALGKK6 pKa = 10.46RR7 pKa = 11.84ILEE10 pKa = 4.1IVSTRR15 pKa = 11.84VEE17 pKa = 4.58DD18 pKa = 4.84DD19 pKa = 4.12DD20 pKa = 4.1GTPAVPLAKK29 pKa = 9.93RR30 pKa = 11.84PRR32 pKa = 11.84CDD34 pKa = 3.34NMPEE38 pKa = 4.18PGCAGIAVGDD48 pKa = 3.55TWNGIEE54 pKa = 4.69CDD56 pKa = 3.81EE57 pKa = 4.74EE58 pKa = 4.51DD59 pKa = 4.41VEE61 pKa = 4.61ARR63 pKa = 11.84DD64 pKa = 4.1HH65 pKa = 6.03IVEE68 pKa = 4.22VVLAGGALATVLDD81 pKa = 4.23QASCHH86 pKa = 6.51HH87 pKa = 7.13IDD89 pKa = 4.07LALIQLFDD97 pKa = 4.43DD98 pKa = 4.06CTALARR104 pKa = 11.84LCRR107 pKa = 11.84AIAPSLPQALPAVLAADD124 pKa = 4.09SEE126 pKa = 4.49PDD128 pKa = 2.93KK129 pKa = 10.91TVYY132 pKa = 10.38HH133 pKa = 5.02VTIDD137 pKa = 3.76ALLCRR142 pKa = 11.84AAQQGRR148 pKa = 11.84PAVLAVLMRR157 pKa = 11.84HH158 pKa = 6.01LADD161 pKa = 4.24EE162 pKa = 4.44YY163 pKa = 11.35DD164 pKa = 3.49VARR167 pKa = 11.84ALEE170 pKa = 4.2MADD173 pKa = 4.7EE174 pKa = 4.71EE175 pKa = 4.97DD176 pKa = 4.27DD177 pKa = 3.86PAAIEE182 pKa = 4.99LIAEE186 pKa = 4.49TYY188 pKa = 10.41EE189 pKa = 4.09KK190 pKa = 10.87DD191 pKa = 3.58AGKK194 pKa = 10.34DD195 pKa = 3.27DD196 pKa = 3.79TGEE199 pKa = 4.72DD200 pKa = 3.67DD201 pKa = 4.04TGEE204 pKa = 4.77DD205 pKa = 3.68DD206 pKa = 3.75TGEE209 pKa = 4.69DD210 pKa = 3.38EE211 pKa = 4.4TGSCRR216 pKa = 11.84RR217 pKa = 11.84IAYY220 pKa = 8.84VALTCAVHH228 pKa = 6.23SSRR231 pKa = 11.84VALIHH236 pKa = 6.86RR237 pKa = 11.84LVDD240 pKa = 3.14KK241 pKa = 11.17CDD243 pKa = 3.4VEE245 pKa = 5.08DD246 pKa = 3.67VHH248 pKa = 8.1GLLDD252 pKa = 4.45DD253 pKa = 4.32CLEE256 pKa = 4.9DD257 pKa = 4.78GDD259 pKa = 4.59VFAALWQHH267 pKa = 6.65AVLCAHH273 pKa = 7.62AYY275 pKa = 8.98AASLPPCPALDD286 pKa = 3.72HH287 pKa = 6.35LTLQIANGEE296 pKa = 4.19PCADD300 pKa = 3.17MCASYY305 pKa = 10.99EE306 pKa = 4.15PADD309 pKa = 4.17SDD311 pKa = 5.13NDD313 pKa = 3.99TDD315 pKa = 6.49DD316 pKa = 5.31DD317 pKa = 4.39TDD319 pKa = 5.77DD320 pKa = 6.2DD321 pKa = 5.5DD322 pKa = 7.18GGDD325 pKa = 4.1NGNDD329 pKa = 4.38DD330 pKa = 5.53DD331 pKa = 6.73DD332 pKa = 6.51NDD334 pKa = 4.33DD335 pKa = 4.17ADD337 pKa = 4.06QGSHH341 pKa = 5.42EE342 pKa = 4.23QAQQ345 pKa = 3.42
Molecular weight: 36.84 kDa
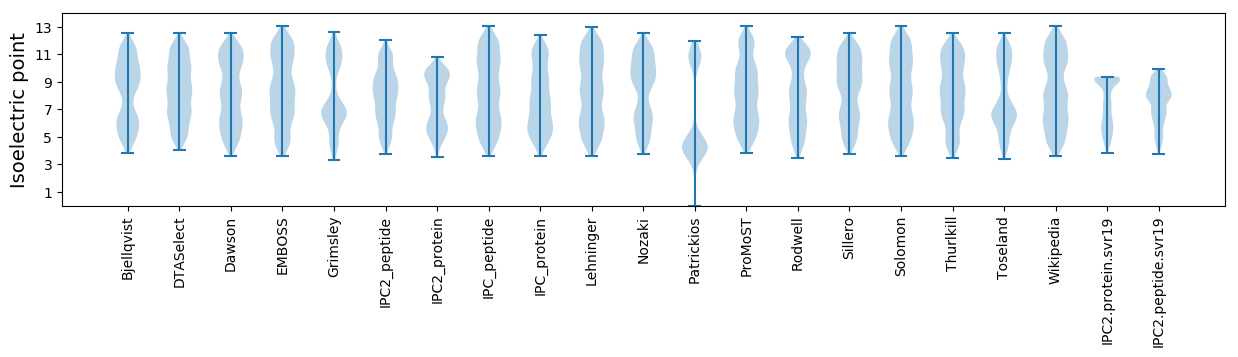
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B5J4K2|A0A0B5J4K2_9VIRU Uncharacterized protein OS=Pandoravirus inopinatum OX=1605721 PE=4 SV=1
MM1 pKa = 7.09TWFFATVAGGGLGVGGRR18 pKa = 11.84AAGVRR23 pKa = 11.84PLCRR27 pKa = 11.84AVILALCRR35 pKa = 11.84QTRR38 pKa = 11.84IRR40 pKa = 11.84RR41 pKa = 11.84VSRR44 pKa = 11.84ADD46 pKa = 3.57FAPSSPLLIFFPIGHH61 pKa = 7.73LRR63 pKa = 11.84RR64 pKa = 11.84QTRR67 pKa = 11.84VWSRR71 pKa = 11.84LSTATVSFPFRR82 pKa = 11.84LFFQTLPFSNWAAVVAHH99 pKa = 6.98APPAEE104 pKa = 4.22ATQKK108 pKa = 8.9VAKK111 pKa = 10.49NFFGRR116 pKa = 11.84FAMIIFPFFFLIGGGAFSWGRR137 pKa = 11.84RR138 pKa = 11.84RR139 pKa = 11.84QKK141 pKa = 8.44STSRR145 pKa = 11.84RR146 pKa = 11.84RR147 pKa = 11.84VLPFAWPTPRR157 pKa = 11.84AAFVPFALQPSKK169 pKa = 10.79INRR172 pKa = 11.84FGAWHH177 pKa = 6.11MSKK180 pKa = 8.5ITSNIANGLPLARR193 pKa = 11.84LKK195 pKa = 10.43AQCRR199 pKa = 11.84QATFFLFKK207 pKa = 9.74WKK209 pKa = 9.73HH210 pKa = 4.24ARR212 pKa = 11.84KK213 pKa = 9.37KK214 pKa = 10.04KK215 pKa = 9.53RR216 pKa = 11.84PARR219 pKa = 11.84RR220 pKa = 11.84RR221 pKa = 11.84LL222 pKa = 3.29
MM1 pKa = 7.09TWFFATVAGGGLGVGGRR18 pKa = 11.84AAGVRR23 pKa = 11.84PLCRR27 pKa = 11.84AVILALCRR35 pKa = 11.84QTRR38 pKa = 11.84IRR40 pKa = 11.84RR41 pKa = 11.84VSRR44 pKa = 11.84ADD46 pKa = 3.57FAPSSPLLIFFPIGHH61 pKa = 7.73LRR63 pKa = 11.84RR64 pKa = 11.84QTRR67 pKa = 11.84VWSRR71 pKa = 11.84LSTATVSFPFRR82 pKa = 11.84LFFQTLPFSNWAAVVAHH99 pKa = 6.98APPAEE104 pKa = 4.22ATQKK108 pKa = 8.9VAKK111 pKa = 10.49NFFGRR116 pKa = 11.84FAMIIFPFFFLIGGGAFSWGRR137 pKa = 11.84RR138 pKa = 11.84RR139 pKa = 11.84QKK141 pKa = 8.44STSRR145 pKa = 11.84RR146 pKa = 11.84RR147 pKa = 11.84VLPFAWPTPRR157 pKa = 11.84AAFVPFALQPSKK169 pKa = 10.79INRR172 pKa = 11.84FGAWHH177 pKa = 6.11MSKK180 pKa = 8.5ITSNIANGLPLARR193 pKa = 11.84LKK195 pKa = 10.43AQCRR199 pKa = 11.84QATFFLFKK207 pKa = 9.74WKK209 pKa = 9.73HH210 pKa = 4.24ARR212 pKa = 11.84KK213 pKa = 9.37KK214 pKa = 10.04KK215 pKa = 9.53RR216 pKa = 11.84PARR219 pKa = 11.84RR220 pKa = 11.84RR221 pKa = 11.84LL222 pKa = 3.29
Molecular weight: 25.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
542761 |
99 |
1504 |
295.1 |
32.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.172 ± 0.083 | 2.948 ± 0.044 |
6.738 ± 0.066 | 3.481 ± 0.039 |
2.969 ± 0.047 | 7.894 ± 0.065 |
3.177 ± 0.039 | 3.154 ± 0.027 |
2.566 ± 0.046 | 8.146 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.056 ± 0.022 | 2.173 ± 0.028 |
6.652 ± 0.054 | 2.736 ± 0.034 |
9.044 ± 0.059 | 6.03 ± 0.057 |
5.852 ± 0.051 | 7.11 ± 0.039 |
2.122 ± 0.036 | 1.978 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
