
Asticcacaulis biprosthecum C19
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Caulobacterales; Caulobacteraceae; Asticcacaulis; Asticcacaulis biprosthecium
Average proteome isoelectric point is 6.4
Get precalculated fractions of proteins
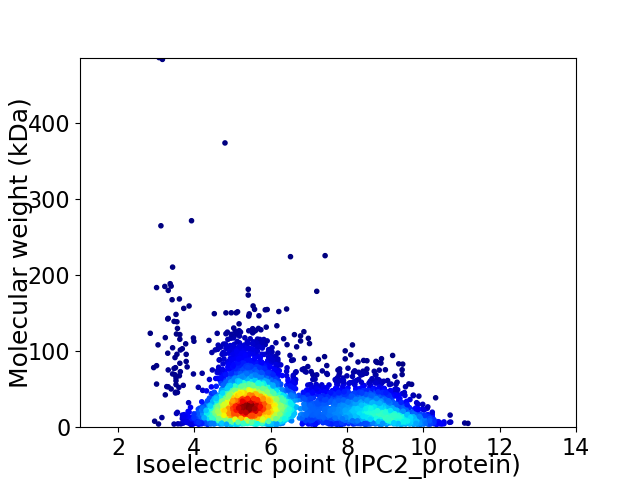
Virtual 2D-PAGE plot for 4712 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
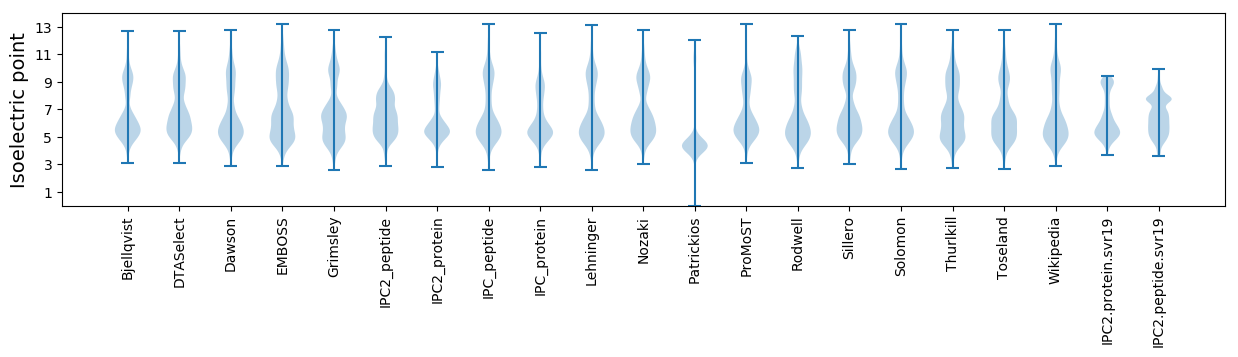
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F4QGL9|F4QGL9_9CAUL Siderophore-interacting family protein OS=Asticcacaulis biprosthecum C19 OX=715226 GN=ABI_21420 PE=4 SV=1
MM1 pKa = 7.89ADD3 pKa = 3.49FTVEE7 pKa = 3.91PVDD10 pKa = 3.84TVNTTTAGTQRR21 pKa = 11.84APEE24 pKa = 3.91ITGLDD29 pKa = 3.09GGGYY33 pKa = 9.29VVTWYY38 pKa = 11.13GNTADD43 pKa = 5.12LGTVLYY49 pKa = 10.07AQVYY53 pKa = 9.13GADD56 pKa = 3.77GDD58 pKa = 4.11KK59 pKa = 11.19LGGEE63 pKa = 4.6VVVADD68 pKa = 4.34HH69 pKa = 6.83LPVTGADD76 pKa = 3.08VAARR80 pKa = 11.84ADD82 pKa = 3.46GGFVVIYY89 pKa = 9.14TEE91 pKa = 4.25AGSAFGIAGDD101 pKa = 3.86PPVIAGRR108 pKa = 11.84HH109 pKa = 4.45FDD111 pKa = 3.41ATGAATSAAYY121 pKa = 9.87AISQAPTLDD130 pKa = 3.6QINPDD135 pKa = 3.23IARR138 pKa = 11.84LADD141 pKa = 3.57GGFMVTWSGQQQASSSDD158 pKa = 3.77HH159 pKa = 6.17NLHH162 pKa = 5.63GQKK165 pKa = 10.62LNLYY169 pKa = 6.84GQRR172 pKa = 11.84VGDD175 pKa = 3.96MQYY178 pKa = 10.75VAANLSYY185 pKa = 10.53QGWAPNPTAGDD196 pKa = 3.4SGTSVIARR204 pKa = 11.84SDD206 pKa = 3.58GGWIVGWMTGRR217 pKa = 11.84SASTAFLTLQAYY229 pKa = 7.34NASGAPVGVAVEE241 pKa = 4.57VKK243 pKa = 9.92PGTYY247 pKa = 9.44PVSSFDD253 pKa = 3.95ADD255 pKa = 3.25ISVLDD260 pKa = 4.38DD261 pKa = 3.36LTVAFNSQSRR271 pKa = 11.84VFIADD276 pKa = 4.25LSTGAVTSFDD286 pKa = 4.4SNVSANTGLLADD298 pKa = 4.08IQGLPDD304 pKa = 3.85GSLLVTWTEE313 pKa = 3.63VDD315 pKa = 3.55GAAGLLKK322 pKa = 10.53AAIYY326 pKa = 10.52DD327 pKa = 3.62RR328 pKa = 11.84AGVVLSDD335 pKa = 3.27IVTVTAAGNNRR346 pKa = 11.84YY347 pKa = 7.97ATATMLSDD355 pKa = 4.13GNVALAWTDD364 pKa = 3.64TVSSDD369 pKa = 3.36IEE371 pKa = 4.15TTIILRR377 pKa = 11.84NHH379 pKa = 5.98GPEE382 pKa = 4.01LTGEE386 pKa = 4.18PVVLPGATEE395 pKa = 3.92DD396 pKa = 3.48TVYY399 pKa = 10.59TLSTADD405 pKa = 4.15LLAGYY410 pKa = 8.12TDD412 pKa = 3.56TDD414 pKa = 3.86GDD416 pKa = 4.24SLSVKK421 pKa = 10.46SLTLEE426 pKa = 4.29GGTLVNNGDD435 pKa = 3.88GTYY438 pKa = 10.52SWTPPANYY446 pKa = 9.42FGPIGLSYY454 pKa = 10.32KK455 pKa = 10.35ISDD458 pKa = 3.72GQGANLIVTRR468 pKa = 11.84SFEE471 pKa = 4.2VARR474 pKa = 11.84VTDD477 pKa = 4.21TIFNSNSTRR486 pKa = 11.84LDD488 pKa = 3.36QYY490 pKa = 11.53SEE492 pKa = 3.9NLTLTGRR499 pKa = 11.84GNIKK503 pKa = 10.52GVGNSWDD510 pKa = 3.55NVLTGNSGINTLEE523 pKa = 4.15GQLGNDD529 pKa = 3.23TYY531 pKa = 11.58YY532 pKa = 10.72VQNSEE537 pKa = 4.09DD538 pKa = 3.97HH539 pKa = 5.66IVEE542 pKa = 4.08YY543 pKa = 11.01RR544 pKa = 11.84NGGVDD549 pKa = 3.77LVYY552 pKa = 11.08ASVDD556 pKa = 3.33YY557 pKa = 11.21SLAGGDD563 pKa = 3.68TEE565 pKa = 5.07YY566 pKa = 11.64LFLTGTADD574 pKa = 3.76LNGRR578 pKa = 11.84GNASANTLAGNGGANRR594 pKa = 11.84LSGDD598 pKa = 3.56RR599 pKa = 11.84GHH601 pKa = 7.44DD602 pKa = 3.48RR603 pKa = 11.84LNGGAGADD611 pKa = 3.74TLIGATGNDD620 pKa = 3.65TYY622 pKa = 11.78VVDD625 pKa = 3.93NVGDD629 pKa = 4.01SVSDD633 pKa = 3.25VDD635 pKa = 4.5EE636 pKa = 4.83NFFSGGTDD644 pKa = 3.17VVEE647 pKa = 5.44ASVSWTLGTDD657 pKa = 3.97LEE659 pKa = 4.35QLVLTGAAAIDD670 pKa = 3.85GTGNGLSNVLTGNDD684 pKa = 3.3ARR686 pKa = 11.84NVLTGGLGNDD696 pKa = 3.27TYY698 pKa = 11.33YY699 pKa = 10.37IQNEE703 pKa = 3.93TDD705 pKa = 3.22RR706 pKa = 11.84VVEE709 pKa = 3.76LHH711 pKa = 6.92FEE713 pKa = 4.39GEE715 pKa = 4.28DD716 pKa = 3.65TVISSVSYY724 pKa = 10.73SLFGRR729 pKa = 11.84AVEE732 pKa = 4.21TLLLTGTGNLNGTGNSLNNTLTGTDD757 pKa = 3.75GNNVLDD763 pKa = 4.67GGTGGDD769 pKa = 4.06LMSGGLGNDD778 pKa = 3.24TYY780 pKa = 11.68YY781 pKa = 11.34VDD783 pKa = 3.96NFYY786 pKa = 11.53DD787 pKa = 3.82NVAEE791 pKa = 4.82LHH793 pKa = 6.09LQGNDD798 pKa = 2.9TVYY801 pKa = 11.51ASVTYY806 pKa = 10.86SFFGRR811 pKa = 11.84AAEE814 pKa = 3.96VLILTGAGNINGTGNSLVNTLIGNSGANVLDD845 pKa = 4.14GAGGNDD851 pKa = 3.68NLTGGAGADD860 pKa = 3.25VFLFSAGSRR869 pKa = 11.84ADD871 pKa = 3.3TVTDD875 pKa = 4.77FNAADD880 pKa = 3.75NDD882 pKa = 4.25SLNVNAYY889 pKa = 8.63TGGVANAGLVSQTGNHH905 pKa = 5.02VVINLGGGNVVTVLNASQAEE925 pKa = 4.35ILAHH929 pKa = 5.2MVWW932 pKa = 3.49
MM1 pKa = 7.89ADD3 pKa = 3.49FTVEE7 pKa = 3.91PVDD10 pKa = 3.84TVNTTTAGTQRR21 pKa = 11.84APEE24 pKa = 3.91ITGLDD29 pKa = 3.09GGGYY33 pKa = 9.29VVTWYY38 pKa = 11.13GNTADD43 pKa = 5.12LGTVLYY49 pKa = 10.07AQVYY53 pKa = 9.13GADD56 pKa = 3.77GDD58 pKa = 4.11KK59 pKa = 11.19LGGEE63 pKa = 4.6VVVADD68 pKa = 4.34HH69 pKa = 6.83LPVTGADD76 pKa = 3.08VAARR80 pKa = 11.84ADD82 pKa = 3.46GGFVVIYY89 pKa = 9.14TEE91 pKa = 4.25AGSAFGIAGDD101 pKa = 3.86PPVIAGRR108 pKa = 11.84HH109 pKa = 4.45FDD111 pKa = 3.41ATGAATSAAYY121 pKa = 9.87AISQAPTLDD130 pKa = 3.6QINPDD135 pKa = 3.23IARR138 pKa = 11.84LADD141 pKa = 3.57GGFMVTWSGQQQASSSDD158 pKa = 3.77HH159 pKa = 6.17NLHH162 pKa = 5.63GQKK165 pKa = 10.62LNLYY169 pKa = 6.84GQRR172 pKa = 11.84VGDD175 pKa = 3.96MQYY178 pKa = 10.75VAANLSYY185 pKa = 10.53QGWAPNPTAGDD196 pKa = 3.4SGTSVIARR204 pKa = 11.84SDD206 pKa = 3.58GGWIVGWMTGRR217 pKa = 11.84SASTAFLTLQAYY229 pKa = 7.34NASGAPVGVAVEE241 pKa = 4.57VKK243 pKa = 9.92PGTYY247 pKa = 9.44PVSSFDD253 pKa = 3.95ADD255 pKa = 3.25ISVLDD260 pKa = 4.38DD261 pKa = 3.36LTVAFNSQSRR271 pKa = 11.84VFIADD276 pKa = 4.25LSTGAVTSFDD286 pKa = 4.4SNVSANTGLLADD298 pKa = 4.08IQGLPDD304 pKa = 3.85GSLLVTWTEE313 pKa = 3.63VDD315 pKa = 3.55GAAGLLKK322 pKa = 10.53AAIYY326 pKa = 10.52DD327 pKa = 3.62RR328 pKa = 11.84AGVVLSDD335 pKa = 3.27IVTVTAAGNNRR346 pKa = 11.84YY347 pKa = 7.97ATATMLSDD355 pKa = 4.13GNVALAWTDD364 pKa = 3.64TVSSDD369 pKa = 3.36IEE371 pKa = 4.15TTIILRR377 pKa = 11.84NHH379 pKa = 5.98GPEE382 pKa = 4.01LTGEE386 pKa = 4.18PVVLPGATEE395 pKa = 3.92DD396 pKa = 3.48TVYY399 pKa = 10.59TLSTADD405 pKa = 4.15LLAGYY410 pKa = 8.12TDD412 pKa = 3.56TDD414 pKa = 3.86GDD416 pKa = 4.24SLSVKK421 pKa = 10.46SLTLEE426 pKa = 4.29GGTLVNNGDD435 pKa = 3.88GTYY438 pKa = 10.52SWTPPANYY446 pKa = 9.42FGPIGLSYY454 pKa = 10.32KK455 pKa = 10.35ISDD458 pKa = 3.72GQGANLIVTRR468 pKa = 11.84SFEE471 pKa = 4.2VARR474 pKa = 11.84VTDD477 pKa = 4.21TIFNSNSTRR486 pKa = 11.84LDD488 pKa = 3.36QYY490 pKa = 11.53SEE492 pKa = 3.9NLTLTGRR499 pKa = 11.84GNIKK503 pKa = 10.52GVGNSWDD510 pKa = 3.55NVLTGNSGINTLEE523 pKa = 4.15GQLGNDD529 pKa = 3.23TYY531 pKa = 11.58YY532 pKa = 10.72VQNSEE537 pKa = 4.09DD538 pKa = 3.97HH539 pKa = 5.66IVEE542 pKa = 4.08YY543 pKa = 11.01RR544 pKa = 11.84NGGVDD549 pKa = 3.77LVYY552 pKa = 11.08ASVDD556 pKa = 3.33YY557 pKa = 11.21SLAGGDD563 pKa = 3.68TEE565 pKa = 5.07YY566 pKa = 11.64LFLTGTADD574 pKa = 3.76LNGRR578 pKa = 11.84GNASANTLAGNGGANRR594 pKa = 11.84LSGDD598 pKa = 3.56RR599 pKa = 11.84GHH601 pKa = 7.44DD602 pKa = 3.48RR603 pKa = 11.84LNGGAGADD611 pKa = 3.74TLIGATGNDD620 pKa = 3.65TYY622 pKa = 11.78VVDD625 pKa = 3.93NVGDD629 pKa = 4.01SVSDD633 pKa = 3.25VDD635 pKa = 4.5EE636 pKa = 4.83NFFSGGTDD644 pKa = 3.17VVEE647 pKa = 5.44ASVSWTLGTDD657 pKa = 3.97LEE659 pKa = 4.35QLVLTGAAAIDD670 pKa = 3.85GTGNGLSNVLTGNDD684 pKa = 3.3ARR686 pKa = 11.84NVLTGGLGNDD696 pKa = 3.27TYY698 pKa = 11.33YY699 pKa = 10.37IQNEE703 pKa = 3.93TDD705 pKa = 3.22RR706 pKa = 11.84VVEE709 pKa = 3.76LHH711 pKa = 6.92FEE713 pKa = 4.39GEE715 pKa = 4.28DD716 pKa = 3.65TVISSVSYY724 pKa = 10.73SLFGRR729 pKa = 11.84AVEE732 pKa = 4.21TLLLTGTGNLNGTGNSLNNTLTGTDD757 pKa = 3.75GNNVLDD763 pKa = 4.67GGTGGDD769 pKa = 4.06LMSGGLGNDD778 pKa = 3.24TYY780 pKa = 11.68YY781 pKa = 11.34VDD783 pKa = 3.96NFYY786 pKa = 11.53DD787 pKa = 3.82NVAEE791 pKa = 4.82LHH793 pKa = 6.09LQGNDD798 pKa = 2.9TVYY801 pKa = 11.51ASVTYY806 pKa = 10.86SFFGRR811 pKa = 11.84AAEE814 pKa = 3.96VLILTGAGNINGTGNSLVNTLIGNSGANVLDD845 pKa = 4.14GAGGNDD851 pKa = 3.68NLTGGAGADD860 pKa = 3.25VFLFSAGSRR869 pKa = 11.84ADD871 pKa = 3.3TVTDD875 pKa = 4.77FNAADD880 pKa = 3.75NDD882 pKa = 4.25SLNVNAYY889 pKa = 8.63TGGVANAGLVSQTGNHH905 pKa = 5.02VVINLGGGNVVTVLNASQAEE925 pKa = 4.35ILAHH929 pKa = 5.2MVWW932 pKa = 3.49
Molecular weight: 95.72 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F4QN39|F4QN39_9CAUL Tad domain-containing protein OS=Asticcacaulis biprosthecum C19 OX=715226 GN=ABI_30470 PE=4 SV=1
MM1 pKa = 8.17RR2 pKa = 11.84YY3 pKa = 10.03LSFLSRR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84FTSRR16 pKa = 11.84ILRR19 pKa = 11.84RR20 pKa = 11.84KK21 pKa = 9.2GLHH24 pKa = 5.46TRR26 pKa = 11.84LILQVGSARR35 pKa = 11.84QRR37 pKa = 11.84RR38 pKa = 11.84KK39 pKa = 9.93RR40 pKa = 3.46
MM1 pKa = 8.17RR2 pKa = 11.84YY3 pKa = 10.03LSFLSRR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84FTSRR16 pKa = 11.84ILRR19 pKa = 11.84RR20 pKa = 11.84KK21 pKa = 9.2GLHH24 pKa = 5.46TRR26 pKa = 11.84LILQVGSARR35 pKa = 11.84QRR37 pKa = 11.84RR38 pKa = 11.84KK39 pKa = 9.93RR40 pKa = 3.46
Molecular weight: 5.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1501214 |
37 |
4917 |
318.6 |
34.49 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.699 ± 0.052 | 0.8 ± 0.013 |
6.362 ± 0.048 | 5.047 ± 0.032 |
3.77 ± 0.026 | 8.87 ± 0.09 |
1.992 ± 0.019 | 4.963 ± 0.022 |
3.703 ± 0.033 | 9.693 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.368 ± 0.019 | 3.244 ± 0.042 |
4.829 ± 0.038 | 3.363 ± 0.023 |
6.103 ± 0.049 | 5.658 ± 0.032 |
5.907 ± 0.042 | 7.315 ± 0.031 |
1.435 ± 0.017 | 2.684 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
