
Human papillomavirus type 88
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Gammapapillomavirus; Gammapapillomavirus 5
Average proteome isoelectric point is 6.3
Get precalculated fractions of proteins
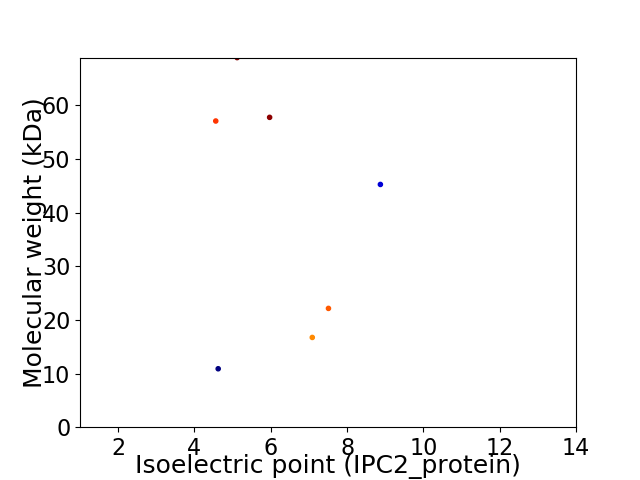
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
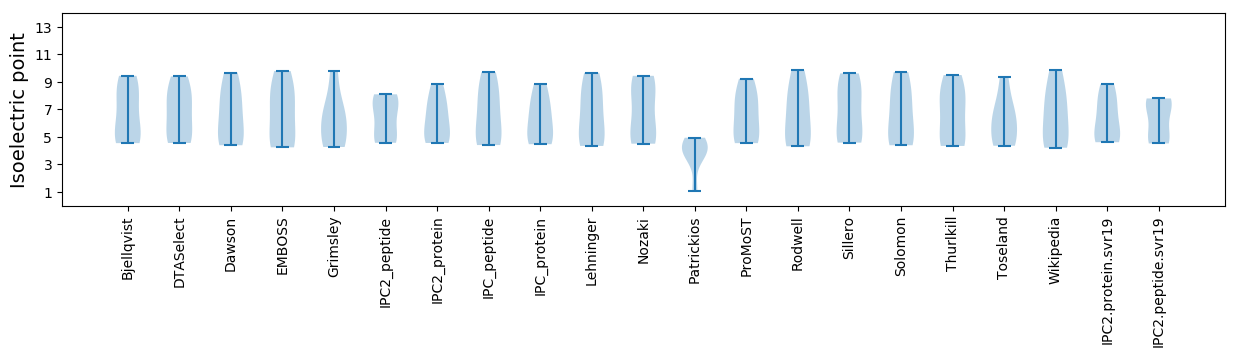
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A8R8N1|A8R8N1_9PAPI Replication protein E1 OS=Human papillomavirus type 88 OX=337054 GN=E1 PE=3 SV=1
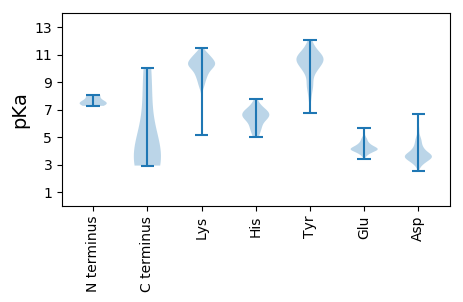
MM1 pKa = 7.45IGKK4 pKa = 9.12EE5 pKa = 3.69PSINDD10 pKa = 3.58LEE12 pKa = 4.63IKK14 pKa = 10.57LEE16 pKa = 4.07EE17 pKa = 4.35HH18 pKa = 6.23VLPANLLSNEE28 pKa = 4.07VLSSDD33 pKa = 4.04EE34 pKa = 3.96EE35 pKa = 4.38TQEE38 pKa = 3.96EE39 pKa = 4.81EE40 pKa = 3.9EE41 pKa = 4.56RR42 pKa = 11.84EE43 pKa = 4.03PFQIDD48 pKa = 3.52TSCAFCEE55 pKa = 3.87AGVRR59 pKa = 11.84VFVLASPAGIRR70 pKa = 11.84TLQQLLLAEE79 pKa = 4.82ISISCPGCSRR89 pKa = 11.84NNFRR93 pKa = 11.84HH94 pKa = 5.91GRR96 pKa = 11.84PQQ98 pKa = 2.92
MM1 pKa = 7.45IGKK4 pKa = 9.12EE5 pKa = 3.69PSINDD10 pKa = 3.58LEE12 pKa = 4.63IKK14 pKa = 10.57LEE16 pKa = 4.07EE17 pKa = 4.35HH18 pKa = 6.23VLPANLLSNEE28 pKa = 4.07VLSSDD33 pKa = 4.04EE34 pKa = 3.96EE35 pKa = 4.38TQEE38 pKa = 3.96EE39 pKa = 4.81EE40 pKa = 3.9EE41 pKa = 4.56RR42 pKa = 11.84EE43 pKa = 4.03PFQIDD48 pKa = 3.52TSCAFCEE55 pKa = 3.87AGVRR59 pKa = 11.84VFVLASPAGIRR70 pKa = 11.84TLQQLLLAEE79 pKa = 4.82ISISCPGCSRR89 pKa = 11.84NNFRR93 pKa = 11.84HH94 pKa = 5.91GRR96 pKa = 11.84PQQ98 pKa = 2.92
Molecular weight: 10.9 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A8R8N4|A8R8N4_9PAPI E4 protein OS=Human papillomavirus type 88 OX=337054 GN=E4 PE=4 SV=1
MM1 pKa = 7.49ARR3 pKa = 11.84MEE5 pKa = 4.24TQEE8 pKa = 4.06TLTEE12 pKa = 4.05RR13 pKa = 11.84FVALQDD19 pKa = 4.49AILNLIEE26 pKa = 5.42RR27 pKa = 11.84GEE29 pKa = 4.14TDD31 pKa = 3.72LRR33 pKa = 11.84SQIQYY38 pKa = 9.21WEE40 pKa = 4.3LVRR43 pKa = 11.84KK44 pKa = 8.35EE45 pKa = 4.0QVILYY50 pKa = 6.74YY51 pKa = 10.56ARR53 pKa = 11.84KK54 pKa = 9.58SGYY57 pKa = 9.69NRR59 pKa = 11.84LGLQPTPAPAVSEE72 pKa = 4.39YY73 pKa = 10.31NAKK76 pKa = 9.73QAIHH80 pKa = 6.27LQLMLKK86 pKa = 10.16SLEE89 pKa = 3.9KK90 pKa = 10.84SKK92 pKa = 10.38FAKK95 pKa = 10.28EE96 pKa = 3.82PWSLTDD102 pKa = 4.63ASAEE106 pKa = 4.05LVNTPPRR113 pKa = 11.84DD114 pKa = 3.68CFKK117 pKa = 11.04KK118 pKa = 10.54GGFTVTVYY126 pKa = 10.67FDD128 pKa = 3.53NDD130 pKa = 3.57RR131 pKa = 11.84EE132 pKa = 4.19NSFPYY137 pKa = 8.68TQWEE141 pKa = 4.23HH142 pKa = 7.75IYY144 pKa = 10.83YY145 pKa = 9.87QDD147 pKa = 5.37QNEE150 pKa = 4.18QWHH153 pKa = 5.97KK154 pKa = 10.31VPGGVDD160 pKa = 3.51HH161 pKa = 7.16NGLYY165 pKa = 10.33YY166 pKa = 10.66DD167 pKa = 4.5EE168 pKa = 5.65EE169 pKa = 4.44NTNEE173 pKa = 3.71RR174 pKa = 11.84VYY176 pKa = 10.98FLLFEE181 pKa = 4.94PEE183 pKa = 4.06SQKK186 pKa = 11.07YY187 pKa = 9.76GSSGQWTVHH196 pKa = 5.21YY197 pKa = 10.91KK198 pKa = 8.78NTTVSASATSSSRR211 pKa = 11.84RR212 pKa = 11.84SSPISTKK219 pKa = 10.08TDD221 pKa = 3.54FDD223 pKa = 3.61ATTAGNTTTSAPQRR237 pKa = 11.84SPRR240 pKa = 11.84KK241 pKa = 8.79RR242 pKa = 11.84LQEE245 pKa = 4.18AVSSTTSPPAHH256 pKa = 6.78NLRR259 pKa = 11.84SPGRR263 pKa = 11.84GRR265 pKa = 11.84GEE267 pKa = 4.13GEE269 pKa = 3.77RR270 pKa = 11.84TSGAKK275 pKa = 9.35RR276 pKa = 11.84RR277 pKa = 11.84RR278 pKa = 11.84TATDD282 pKa = 3.21GNTLGEE288 pKa = 4.54SVPSPSQVGSRR299 pKa = 11.84HH300 pKa = 5.66RR301 pKa = 11.84APEE304 pKa = 3.74RR305 pKa = 11.84SGLSRR310 pKa = 11.84LGRR313 pKa = 11.84LQADD317 pKa = 3.04AWDD320 pKa = 4.26PPLIIIKK327 pKa = 10.59GPANTLKK334 pKa = 10.47CWRR337 pKa = 11.84NRR339 pKa = 11.84MKK341 pKa = 10.82KK342 pKa = 9.84NSSSNLVCSSVWRR355 pKa = 11.84WIDD358 pKa = 3.21STTHH362 pKa = 5.49EE363 pKa = 4.27NSRR366 pKa = 11.84MLVAFQNTAEE376 pKa = 4.22RR377 pKa = 11.84TRR379 pKa = 11.84FLNSVTLPKK388 pKa = 10.03GTTYY392 pKa = 11.46AFGYY396 pKa = 10.35LDD398 pKa = 3.51SLL400 pKa = 4.27
MM1 pKa = 7.49ARR3 pKa = 11.84MEE5 pKa = 4.24TQEE8 pKa = 4.06TLTEE12 pKa = 4.05RR13 pKa = 11.84FVALQDD19 pKa = 4.49AILNLIEE26 pKa = 5.42RR27 pKa = 11.84GEE29 pKa = 4.14TDD31 pKa = 3.72LRR33 pKa = 11.84SQIQYY38 pKa = 9.21WEE40 pKa = 4.3LVRR43 pKa = 11.84KK44 pKa = 8.35EE45 pKa = 4.0QVILYY50 pKa = 6.74YY51 pKa = 10.56ARR53 pKa = 11.84KK54 pKa = 9.58SGYY57 pKa = 9.69NRR59 pKa = 11.84LGLQPTPAPAVSEE72 pKa = 4.39YY73 pKa = 10.31NAKK76 pKa = 9.73QAIHH80 pKa = 6.27LQLMLKK86 pKa = 10.16SLEE89 pKa = 3.9KK90 pKa = 10.84SKK92 pKa = 10.38FAKK95 pKa = 10.28EE96 pKa = 3.82PWSLTDD102 pKa = 4.63ASAEE106 pKa = 4.05LVNTPPRR113 pKa = 11.84DD114 pKa = 3.68CFKK117 pKa = 11.04KK118 pKa = 10.54GGFTVTVYY126 pKa = 10.67FDD128 pKa = 3.53NDD130 pKa = 3.57RR131 pKa = 11.84EE132 pKa = 4.19NSFPYY137 pKa = 8.68TQWEE141 pKa = 4.23HH142 pKa = 7.75IYY144 pKa = 10.83YY145 pKa = 9.87QDD147 pKa = 5.37QNEE150 pKa = 4.18QWHH153 pKa = 5.97KK154 pKa = 10.31VPGGVDD160 pKa = 3.51HH161 pKa = 7.16NGLYY165 pKa = 10.33YY166 pKa = 10.66DD167 pKa = 4.5EE168 pKa = 5.65EE169 pKa = 4.44NTNEE173 pKa = 3.71RR174 pKa = 11.84VYY176 pKa = 10.98FLLFEE181 pKa = 4.94PEE183 pKa = 4.06SQKK186 pKa = 11.07YY187 pKa = 9.76GSSGQWTVHH196 pKa = 5.21YY197 pKa = 10.91KK198 pKa = 8.78NTTVSASATSSSRR211 pKa = 11.84RR212 pKa = 11.84SSPISTKK219 pKa = 10.08TDD221 pKa = 3.54FDD223 pKa = 3.61ATTAGNTTTSAPQRR237 pKa = 11.84SPRR240 pKa = 11.84KK241 pKa = 8.79RR242 pKa = 11.84LQEE245 pKa = 4.18AVSSTTSPPAHH256 pKa = 6.78NLRR259 pKa = 11.84SPGRR263 pKa = 11.84GRR265 pKa = 11.84GEE267 pKa = 4.13GEE269 pKa = 3.77RR270 pKa = 11.84TSGAKK275 pKa = 9.35RR276 pKa = 11.84RR277 pKa = 11.84RR278 pKa = 11.84TATDD282 pKa = 3.21GNTLGEE288 pKa = 4.54SVPSPSQVGSRR299 pKa = 11.84HH300 pKa = 5.66RR301 pKa = 11.84APEE304 pKa = 3.74RR305 pKa = 11.84SGLSRR310 pKa = 11.84LGRR313 pKa = 11.84LQADD317 pKa = 3.04AWDD320 pKa = 4.26PPLIIIKK327 pKa = 10.59GPANTLKK334 pKa = 10.47CWRR337 pKa = 11.84NRR339 pKa = 11.84MKK341 pKa = 10.82KK342 pKa = 9.84NSSSNLVCSSVWRR355 pKa = 11.84WIDD358 pKa = 3.21STTHH362 pKa = 5.49EE363 pKa = 4.27NSRR366 pKa = 11.84MLVAFQNTAEE376 pKa = 4.22RR377 pKa = 11.84TRR379 pKa = 11.84FLNSVTLPKK388 pKa = 10.03GTTYY392 pKa = 11.46AFGYY396 pKa = 10.35LDD398 pKa = 3.51SLL400 pKa = 4.27
Molecular weight: 45.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2472 |
98 |
607 |
353.1 |
39.83 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.866 ± 0.382 | 2.225 ± 0.837 |
6.189 ± 0.514 | 6.756 ± 0.761 |
4.854 ± 0.535 | 5.663 ± 0.636 |
1.861 ± 0.086 | 5.461 ± 1.071 |
4.733 ± 0.685 | 9.223 ± 0.781 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.659 ± 0.318 | 5.299 ± 0.5 |
6.675 ± 1.207 | 3.883 ± 0.688 |
6.23 ± 0.757 | 7.241 ± 0.761 |
6.472 ± 0.836 | 5.542 ± 0.503 |
1.173 ± 0.281 | 2.994 ± 0.361 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
