
Cyberlindnera fabianii (Yeast) (Hansenula fabianii)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota;
Average proteome isoelectric point is 6.25
Get precalculated fractions of proteins
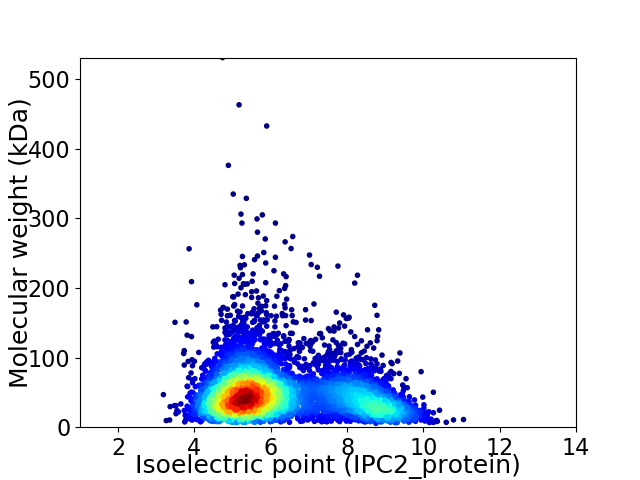
Virtual 2D-PAGE plot for 5507 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A061AVQ0|A0A061AVQ0_CYBFA Casein kinase II subunit beta OS=Cyberlindnera fabianii OX=36022 GN=BON22_3797 PE=3 SV=1
MM1 pKa = 7.52RR2 pKa = 11.84TTTTTNLLASLAWLATTVIAADD24 pKa = 4.15LTPVADD30 pKa = 4.38LFYY33 pKa = 11.02SRR35 pKa = 11.84TSDD38 pKa = 3.34FPLAVEE44 pKa = 4.73LKK46 pKa = 10.65VGSPQQKK53 pKa = 10.19LLAQLDD59 pKa = 4.13TQSRR63 pKa = 11.84DD64 pKa = 3.01ILLSTEE70 pKa = 3.75YY71 pKa = 10.69DD72 pKa = 3.33AVSSIFDD79 pKa = 3.63TNSFNFNADD88 pKa = 3.13SSSSFSTFSGTSWGSDD104 pKa = 2.94NIGFRR109 pKa = 11.84GSDD112 pKa = 2.86ATYY115 pKa = 10.44QDD117 pKa = 3.55VLFGLRR123 pKa = 11.84ASEE126 pKa = 4.32SVPSVFSLGHH136 pKa = 5.19PNIYY140 pKa = 10.38KK141 pKa = 10.07DD142 pKa = 4.25DD143 pKa = 4.39KK144 pKa = 11.39NSFWDD149 pKa = 3.91ALASQNITKK158 pKa = 10.23SYY160 pKa = 10.92SIGFADD166 pKa = 4.91APEE169 pKa = 4.68DD170 pKa = 3.75APGVYY175 pKa = 9.89SAEE178 pKa = 4.25DD179 pKa = 3.25AATVSGLIVFGGVAQAYY196 pKa = 8.29YY197 pKa = 10.97NSLTILPLASNDD209 pKa = 3.6DD210 pKa = 4.5LIGFTLTAVGVQADD224 pKa = 4.39TEE226 pKa = 4.51SSPKK230 pKa = 9.37TISAIKK236 pKa = 9.74HH237 pKa = 4.84LAVVKK242 pKa = 9.83TRR244 pKa = 11.84GIFPSMPKK252 pKa = 9.94QLVEE256 pKa = 4.54NIVTSLDD263 pKa = 3.32KK264 pKa = 10.72DD265 pKa = 3.92TEE267 pKa = 4.43LTTGTSGLYY276 pKa = 10.0EE277 pKa = 4.13VDD279 pKa = 3.89CDD281 pKa = 4.48TEE283 pKa = 4.36FTLLFNFQGAVIRR296 pKa = 11.84IPSTEE301 pKa = 4.62LLTEE305 pKa = 4.33SDD307 pKa = 5.04DD308 pKa = 4.4GSCSLAIIPSDD319 pKa = 3.19SDD321 pKa = 3.41EE322 pKa = 4.12FVIAGGMLTYY332 pKa = 9.8FYY334 pKa = 10.88FVVDD338 pKa = 4.44FEE340 pKa = 4.87NDD342 pKa = 3.28QISIGQSKK350 pKa = 9.92FPGANVDD357 pKa = 4.18PEE359 pKa = 4.05IEE361 pKa = 4.39TYY363 pKa = 10.33SGSVSGAKK371 pKa = 8.28TAAYY375 pKa = 9.97YY376 pKa = 10.78SEE378 pKa = 4.54TFSSQLPEE386 pKa = 4.04PTSDD390 pKa = 3.6SSLTSEE396 pKa = 4.2ATTDD400 pKa = 3.34SSTHH404 pKa = 6.75DD405 pKa = 3.81SSSADD410 pKa = 3.3QTVTPSSSEE419 pKa = 3.82SSLTITGYY427 pKa = 8.36NTYY430 pKa = 9.85PYY432 pKa = 9.4TNTSSSSSSSSSSSLASSTSTSTSTVEE459 pKa = 3.82APTISGSCINGQLQWTVNIPASIGPFNAFSYY490 pKa = 10.25EE491 pKa = 4.19GTGEE495 pKa = 4.61GYY497 pKa = 9.23TITNVKK503 pKa = 10.42LNGRR507 pKa = 11.84DD508 pKa = 3.45LTSEE512 pKa = 3.98SGINGAGFSLDD523 pKa = 3.27TTDD526 pKa = 2.86INGRR530 pKa = 11.84SEE532 pKa = 4.45SLTLVYY538 pKa = 9.43TAIRR542 pKa = 11.84FDD544 pKa = 3.68STTLFSSDD552 pKa = 2.53GTLAITRR559 pKa = 11.84PNRR562 pKa = 11.84KK563 pKa = 9.02RR564 pKa = 11.84EE565 pKa = 3.8VLIYY569 pKa = 9.58EE570 pKa = 4.31LSYY573 pKa = 11.02TIDD576 pKa = 3.22TALGYY581 pKa = 10.75AVTNSDD587 pKa = 3.57VEE589 pKa = 4.55EE590 pKa = 4.25TSSVATSTDD599 pKa = 3.19VTSVLSTTVVTATSCSMGICIEE621 pKa = 4.14APITTGVTVATSTVHH636 pKa = 5.05GTVTSFTTYY645 pKa = 10.45CPLSTEE651 pKa = 4.33SFTNSTTPIVTPSTVSLSSTFLVTSPFSPSTEE683 pKa = 3.92VGTTSTPVINPQLEE697 pKa = 4.68VQSSPALSSSSITAPSILTYY717 pKa = 10.68SGGAVGSFTSITNLLFVFPLLLLII741 pKa = 5.19
MM1 pKa = 7.52RR2 pKa = 11.84TTTTTNLLASLAWLATTVIAADD24 pKa = 4.15LTPVADD30 pKa = 4.38LFYY33 pKa = 11.02SRR35 pKa = 11.84TSDD38 pKa = 3.34FPLAVEE44 pKa = 4.73LKK46 pKa = 10.65VGSPQQKK53 pKa = 10.19LLAQLDD59 pKa = 4.13TQSRR63 pKa = 11.84DD64 pKa = 3.01ILLSTEE70 pKa = 3.75YY71 pKa = 10.69DD72 pKa = 3.33AVSSIFDD79 pKa = 3.63TNSFNFNADD88 pKa = 3.13SSSSFSTFSGTSWGSDD104 pKa = 2.94NIGFRR109 pKa = 11.84GSDD112 pKa = 2.86ATYY115 pKa = 10.44QDD117 pKa = 3.55VLFGLRR123 pKa = 11.84ASEE126 pKa = 4.32SVPSVFSLGHH136 pKa = 5.19PNIYY140 pKa = 10.38KK141 pKa = 10.07DD142 pKa = 4.25DD143 pKa = 4.39KK144 pKa = 11.39NSFWDD149 pKa = 3.91ALASQNITKK158 pKa = 10.23SYY160 pKa = 10.92SIGFADD166 pKa = 4.91APEE169 pKa = 4.68DD170 pKa = 3.75APGVYY175 pKa = 9.89SAEE178 pKa = 4.25DD179 pKa = 3.25AATVSGLIVFGGVAQAYY196 pKa = 8.29YY197 pKa = 10.97NSLTILPLASNDD209 pKa = 3.6DD210 pKa = 4.5LIGFTLTAVGVQADD224 pKa = 4.39TEE226 pKa = 4.51SSPKK230 pKa = 9.37TISAIKK236 pKa = 9.74HH237 pKa = 4.84LAVVKK242 pKa = 9.83TRR244 pKa = 11.84GIFPSMPKK252 pKa = 9.94QLVEE256 pKa = 4.54NIVTSLDD263 pKa = 3.32KK264 pKa = 10.72DD265 pKa = 3.92TEE267 pKa = 4.43LTTGTSGLYY276 pKa = 10.0EE277 pKa = 4.13VDD279 pKa = 3.89CDD281 pKa = 4.48TEE283 pKa = 4.36FTLLFNFQGAVIRR296 pKa = 11.84IPSTEE301 pKa = 4.62LLTEE305 pKa = 4.33SDD307 pKa = 5.04DD308 pKa = 4.4GSCSLAIIPSDD319 pKa = 3.19SDD321 pKa = 3.41EE322 pKa = 4.12FVIAGGMLTYY332 pKa = 9.8FYY334 pKa = 10.88FVVDD338 pKa = 4.44FEE340 pKa = 4.87NDD342 pKa = 3.28QISIGQSKK350 pKa = 9.92FPGANVDD357 pKa = 4.18PEE359 pKa = 4.05IEE361 pKa = 4.39TYY363 pKa = 10.33SGSVSGAKK371 pKa = 8.28TAAYY375 pKa = 9.97YY376 pKa = 10.78SEE378 pKa = 4.54TFSSQLPEE386 pKa = 4.04PTSDD390 pKa = 3.6SSLTSEE396 pKa = 4.2ATTDD400 pKa = 3.34SSTHH404 pKa = 6.75DD405 pKa = 3.81SSSADD410 pKa = 3.3QTVTPSSSEE419 pKa = 3.82SSLTITGYY427 pKa = 8.36NTYY430 pKa = 9.85PYY432 pKa = 9.4TNTSSSSSSSSSSSLASSTSTSTSTVEE459 pKa = 3.82APTISGSCINGQLQWTVNIPASIGPFNAFSYY490 pKa = 10.25EE491 pKa = 4.19GTGEE495 pKa = 4.61GYY497 pKa = 9.23TITNVKK503 pKa = 10.42LNGRR507 pKa = 11.84DD508 pKa = 3.45LTSEE512 pKa = 3.98SGINGAGFSLDD523 pKa = 3.27TTDD526 pKa = 2.86INGRR530 pKa = 11.84SEE532 pKa = 4.45SLTLVYY538 pKa = 9.43TAIRR542 pKa = 11.84FDD544 pKa = 3.68STTLFSSDD552 pKa = 2.53GTLAITRR559 pKa = 11.84PNRR562 pKa = 11.84KK563 pKa = 9.02RR564 pKa = 11.84EE565 pKa = 3.8VLIYY569 pKa = 9.58EE570 pKa = 4.31LSYY573 pKa = 11.02TIDD576 pKa = 3.22TALGYY581 pKa = 10.75AVTNSDD587 pKa = 3.57VEE589 pKa = 4.55EE590 pKa = 4.25TSSVATSTDD599 pKa = 3.19VTSVLSTTVVTATSCSMGICIEE621 pKa = 4.14APITTGVTVATSTVHH636 pKa = 5.05GTVTSFTTYY645 pKa = 10.45CPLSTEE651 pKa = 4.33SFTNSTTPIVTPSTVSLSSTFLVTSPFSPSTEE683 pKa = 3.92VGTTSTPVINPQLEE697 pKa = 4.68VQSSPALSSSSITAPSILTYY717 pKa = 10.68SGGAVGSFTSITNLLFVFPLLLLII741 pKa = 5.19
Molecular weight: 77.94 kDa
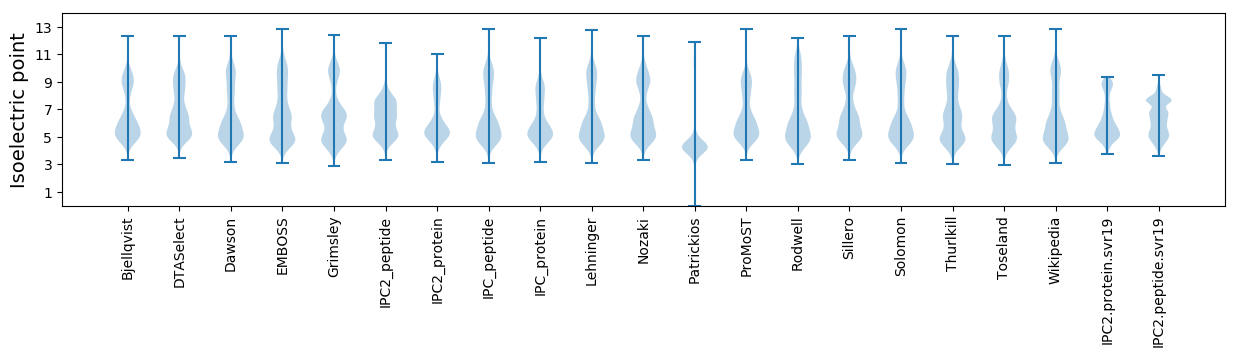
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A061B0L3|A0A061B0L3_CYBFA Nicotinamide-nucleotide adenylyltransferase OS=Cyberlindnera fabianii OX=36022 GN=BON22_3066 PE=4 SV=1
MM1 pKa = 6.68VKK3 pKa = 10.16EE4 pKa = 4.38RR5 pKa = 11.84IEE7 pKa = 4.17VMSRR11 pKa = 11.84LYY13 pKa = 11.0FNFEE17 pKa = 4.0LFRR20 pKa = 11.84ALTFATGKK28 pKa = 10.42RR29 pKa = 11.84IALPTRR35 pKa = 11.84VEE37 pKa = 4.05PKK39 pKa = 10.45VFFANEE45 pKa = 3.78RR46 pKa = 11.84TFLSWLNFTVVLGGLGVGLLNFGDD70 pKa = 3.55KK71 pKa = 10.34VGRR74 pKa = 11.84VSAGLFTFVAMATMVYY90 pKa = 11.12ALITYY95 pKa = 8.06HH96 pKa = 5.58WRR98 pKa = 11.84AVAIRR103 pKa = 11.84KK104 pKa = 9.53RR105 pKa = 11.84GSGPYY110 pKa = 9.62DD111 pKa = 3.48DD112 pKa = 5.8RR113 pKa = 11.84FGPTMLCLFLLVAVVVNFVLRR134 pKa = 11.84IRR136 pKa = 11.84AAA138 pKa = 3.32
MM1 pKa = 6.68VKK3 pKa = 10.16EE4 pKa = 4.38RR5 pKa = 11.84IEE7 pKa = 4.17VMSRR11 pKa = 11.84LYY13 pKa = 11.0FNFEE17 pKa = 4.0LFRR20 pKa = 11.84ALTFATGKK28 pKa = 10.42RR29 pKa = 11.84IALPTRR35 pKa = 11.84VEE37 pKa = 4.05PKK39 pKa = 10.45VFFANEE45 pKa = 3.78RR46 pKa = 11.84TFLSWLNFTVVLGGLGVGLLNFGDD70 pKa = 3.55KK71 pKa = 10.34VGRR74 pKa = 11.84VSAGLFTFVAMATMVYY90 pKa = 11.12ALITYY95 pKa = 8.06HH96 pKa = 5.58WRR98 pKa = 11.84AVAIRR103 pKa = 11.84KK104 pKa = 9.53RR105 pKa = 11.84GSGPYY110 pKa = 9.62DD111 pKa = 3.48DD112 pKa = 5.8RR113 pKa = 11.84FGPTMLCLFLLVAVVVNFVLRR134 pKa = 11.84IRR136 pKa = 11.84AAA138 pKa = 3.32
Molecular weight: 15.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2608749 |
49 |
4725 |
473.7 |
53.09 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.659 ± 0.027 | 1.031 ± 0.01 |
6.137 ± 0.026 | 6.67 ± 0.037 |
4.299 ± 0.023 | 5.556 ± 0.029 |
2.202 ± 0.013 | 5.965 ± 0.023 |
6.778 ± 0.035 | 9.388 ± 0.032 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.075 ± 0.012 | 4.608 ± 0.02 |
4.678 ± 0.029 | 3.968 ± 0.028 |
4.513 ± 0.022 | 8.548 ± 0.048 |
6.325 ± 0.028 | 6.323 ± 0.024 |
1.075 ± 0.011 | 3.203 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
