
Thermoflexibacter ruber
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Cytophagia; Cytophagales;
Average proteome isoelectric point is 6.85
Get precalculated fractions of proteins
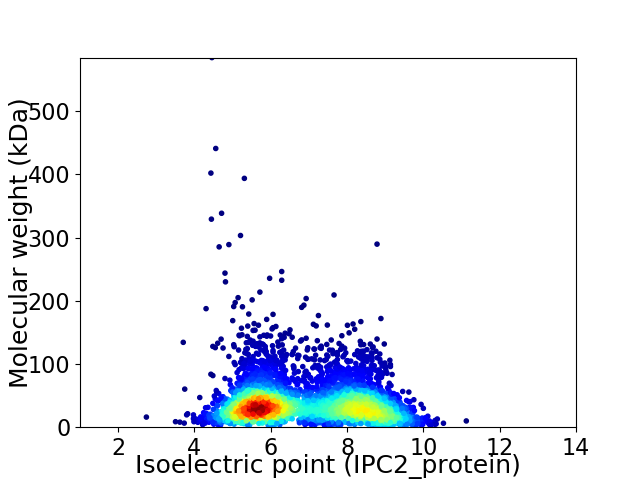
Virtual 2D-PAGE plot for 4522 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
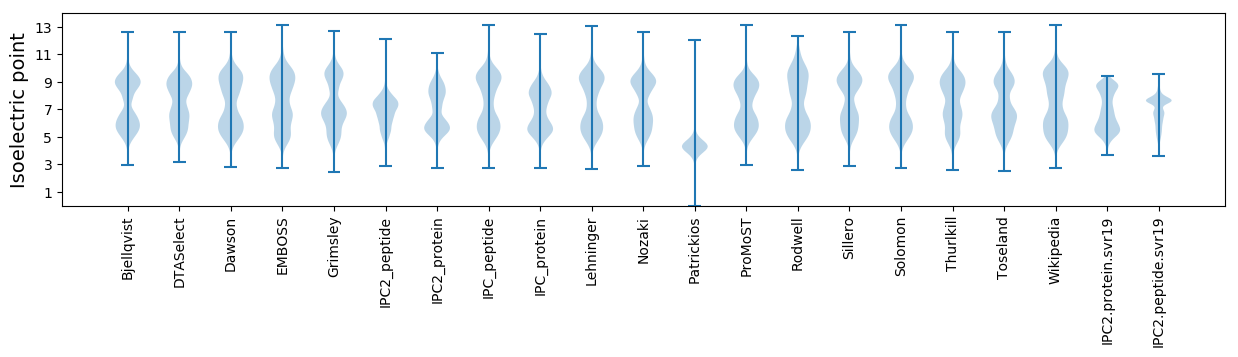
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I2FW50|A0A1I2FW50_9BACT Uncharacterized protein OS=Thermoflexibacter ruber OX=1003 GN=SAMN04488541_1015110 PE=4 SV=1
AA1 pKa = 7.83INPNTQWYY9 pKa = 9.92LDD11 pKa = 3.68NDD13 pKa = 3.51NDD15 pKa = 4.14GFGAGAVAFTQCTKK29 pKa = 9.5PTGNYY34 pKa = 5.87VTNNTDD40 pKa = 3.87CNDD43 pKa = 3.23NDD45 pKa = 3.85TTLNPNTKK53 pKa = 8.86WYY55 pKa = 10.44RR56 pKa = 11.84DD57 pKa = 3.26ADD59 pKa = 3.58GDD61 pKa = 4.59GFGNPADD68 pKa = 4.76EE69 pKa = 4.85KK70 pKa = 9.53TQCLQPAGYY79 pKa = 9.81VRR81 pKa = 11.84NNTDD85 pKa = 3.22CDD87 pKa = 4.24DD88 pKa = 4.15NNVAINPTTVWYY100 pKa = 10.42RR101 pKa = 11.84DD102 pKa = 3.11ADD104 pKa = 3.58GDD106 pKa = 4.44GFGNPAITQVSCTQPTGFVLNNADD130 pKa = 4.77CNDD133 pKa = 3.37SDD135 pKa = 4.38ATLNPNTKK143 pKa = 8.87WYY145 pKa = 10.44RR146 pKa = 11.84DD147 pKa = 3.26ADD149 pKa = 3.58GDD151 pKa = 4.59GFGNPADD158 pKa = 4.76EE159 pKa = 4.85KK160 pKa = 9.53TQCLQPAGYY169 pKa = 9.81VRR171 pKa = 11.84NNTDD175 pKa = 3.43CNDD178 pKa = 3.3NNVAINPTTVWYY190 pKa = 10.39RR191 pKa = 11.84DD192 pKa = 3.17TDD194 pKa = 3.44GDD196 pKa = 4.27GFGNPAITQVSCTQPTGFVLNNADD220 pKa = 4.76CDD222 pKa = 3.94DD223 pKa = 4.08TKK225 pKa = 11.9ADD227 pKa = 3.94INPNTKK233 pKa = 8.89WYY235 pKa = 10.39RR236 pKa = 11.84DD237 pKa = 3.26ADD239 pKa = 3.58GDD241 pKa = 4.59GFGNPADD248 pKa = 4.76EE249 pKa = 4.85KK250 pKa = 9.53TQCLQPAGYY259 pKa = 9.81VRR261 pKa = 11.84NNTDD265 pKa = 3.82CNDD268 pKa = 3.13NNAAINPTTVWYY280 pKa = 10.42RR281 pKa = 11.84DD282 pKa = 3.11ADD284 pKa = 3.58GDD286 pKa = 4.44GFGNPAITQVSCTQPTGFVLNNADD310 pKa = 4.77CNDD313 pKa = 3.37SDD315 pKa = 4.38ATLNPNTKK323 pKa = 8.87WYY325 pKa = 10.44RR326 pKa = 11.84DD327 pKa = 3.26ADD329 pKa = 3.58GDD331 pKa = 4.59GFGNPADD338 pKa = 4.76EE339 pKa = 4.85KK340 pKa = 9.53TQCLQPAGYY349 pKa = 9.81VRR351 pKa = 11.84NNTDD355 pKa = 4.04CDD357 pKa = 3.69DD358 pKa = 3.92TKK360 pKa = 11.9ADD362 pKa = 4.3INPNTQWYY370 pKa = 9.88RR371 pKa = 11.84DD372 pKa = 3.41ADD374 pKa = 3.62GDD376 pKa = 4.59GFGNPADD383 pKa = 4.27VKK385 pKa = 8.09TQCLQPAGYY394 pKa = 9.81VRR396 pKa = 11.84NNTDD400 pKa = 3.43CNDD403 pKa = 3.3NNVAINPTTVWYY415 pKa = 10.42RR416 pKa = 11.84DD417 pKa = 3.11ADD419 pKa = 3.58GDD421 pKa = 4.27GFGNPTITQVSCTQPTGFVLSNADD445 pKa = 4.35CNDD448 pKa = 2.93SDD450 pKa = 4.38ATLNPNTKK458 pKa = 8.87WYY460 pKa = 10.44RR461 pKa = 11.84DD462 pKa = 3.26ADD464 pKa = 3.58GDD466 pKa = 4.59GFGNPADD473 pKa = 5.01AKK475 pKa = 7.57TQCLQPTGYY484 pKa = 10.02IRR486 pKa = 11.84DD487 pKa = 3.8NTDD490 pKa = 3.24CDD492 pKa = 3.7DD493 pKa = 3.87TKK495 pKa = 11.9ADD497 pKa = 4.3INPNTQWYY505 pKa = 9.88RR506 pKa = 11.84DD507 pKa = 3.41ADD509 pKa = 3.62GDD511 pKa = 4.59GFGNPADD518 pKa = 4.26VKK520 pKa = 10.36NQCLQPTGYY529 pKa = 10.47VRR531 pKa = 11.84DD532 pKa = 3.9NTDD535 pKa = 3.72CNDD538 pKa = 3.46SNASLNPNTKK548 pKa = 8.87WYY550 pKa = 10.55ADD552 pKa = 3.51NDD554 pKa = 3.86GDD556 pKa = 4.3GFGAATVAFTQCNKK570 pKa = 9.7PAGNYY575 pKa = 6.2VTNNTDD581 pKa = 4.0CDD583 pKa = 3.8DD584 pKa = 3.92TKK586 pKa = 11.9ADD588 pKa = 3.94INPNTKK594 pKa = 8.89WYY596 pKa = 10.39RR597 pKa = 11.84DD598 pKa = 3.26ADD600 pKa = 3.58GDD602 pKa = 4.74GFGNPANVKK611 pKa = 6.75TQCLQPAGYY620 pKa = 9.95IRR622 pKa = 11.84DD623 pKa = 3.78NTDD626 pKa = 3.55CNDD629 pKa = 3.35SDD631 pKa = 3.85ATLNPNTKK639 pKa = 8.89WYY641 pKa = 10.55ADD643 pKa = 3.51NDD645 pKa = 3.87GDD647 pKa = 4.48GFGAGPVVFVQCAKK661 pKa = 9.28PTGSYY666 pKa = 7.63VTNNGDD672 pKa = 4.62CDD674 pKa = 3.91DD675 pKa = 4.52TNPAIKK681 pKa = 8.61PTAVWYY687 pKa = 10.46RR688 pKa = 11.84DD689 pKa = 3.12ADD691 pKa = 3.56GDD693 pKa = 4.33GFGNPSISVVACSQPAGYY711 pKa = 10.95VMNNTDD717 pKa = 4.82CNDD720 pKa = 3.19NDD722 pKa = 4.34ASINPNTKK730 pKa = 8.92WYY732 pKa = 10.54ADD734 pKa = 3.58NDD736 pKa = 3.62NDD738 pKa = 4.35GFGAGAVAFTQCAKK752 pKa = 10.4PVGNFVTNNSDD763 pKa = 4.61CNDD766 pKa = 3.23NDD768 pKa = 3.38ATLNPNTKK776 pKa = 8.87WYY778 pKa = 10.44RR779 pKa = 11.84DD780 pKa = 3.26ADD782 pKa = 3.58GDD784 pKa = 4.59GFGNPADD791 pKa = 4.27VKK793 pKa = 8.09TQCLQPAGYY802 pKa = 9.95IRR804 pKa = 11.84DD805 pKa = 3.81NTDD808 pKa = 3.24CDD810 pKa = 3.7DD811 pKa = 3.87TKK813 pKa = 11.9ADD815 pKa = 3.94INPNTKK821 pKa = 8.89WYY823 pKa = 10.39RR824 pKa = 11.84DD825 pKa = 3.26ADD827 pKa = 3.58GDD829 pKa = 4.59GFGNPADD836 pKa = 4.06VRR838 pKa = 11.84TQCLQPVGYY847 pKa = 9.96VRR849 pKa = 11.84SNADD853 pKa = 3.9CNDD856 pKa = 3.06SDD858 pKa = 4.38ATLNPNTIWYY868 pKa = 9.54RR869 pKa = 11.84DD870 pKa = 3.26ADD872 pKa = 3.6GDD874 pKa = 4.28GFGNPSVTQKK884 pKa = 10.88SCTQPAGFVRR894 pKa = 11.84NNQDD898 pKa = 3.94CNDD901 pKa = 3.9SDD903 pKa = 5.16ASLNPNTKK911 pKa = 8.84WYY913 pKa = 10.44RR914 pKa = 11.84DD915 pKa = 3.26ADD917 pKa = 3.58GDD919 pKa = 4.59GFGNPADD926 pKa = 4.06VRR928 pKa = 11.84TQCLQPAGYY937 pKa = 9.81VRR939 pKa = 11.84NNTDD943 pKa = 3.72CDD945 pKa = 3.61DD946 pKa = 3.77TNRR949 pKa = 11.84NINPNTKK956 pKa = 9.24WYY958 pKa = 10.43LDD960 pKa = 3.43QDD962 pKa = 3.56GDD964 pKa = 4.86GFGDD968 pKa = 3.58PNNFIVSCTKK978 pKa = 10.27PVGNYY983 pKa = 8.99VLNGTDD989 pKa = 3.42NCPTLPNPNQVIPTWYY1005 pKa = 10.18LDD1007 pKa = 3.31QDD1009 pKa = 3.56RR1010 pKa = 11.84DD1011 pKa = 4.3GLGDD1015 pKa = 3.63PANAIQACSRR1025 pKa = 11.84PDD1027 pKa = 3.43GYY1029 pKa = 11.61VSNNTDD1035 pKa = 3.0NCPTIFNPDD1044 pKa = 2.59GTVPKK1049 pKa = 9.65WYY1051 pKa = 9.87RR1052 pKa = 11.84DD1053 pKa = 3.19ADD1055 pKa = 3.59GDD1057 pKa = 4.39GFGNPNVVVSACTAPIGYY1075 pKa = 9.52VPDD1078 pKa = 3.77NTDD1081 pKa = 4.68CDD1083 pKa = 4.48DD1084 pKa = 4.48SDD1086 pKa = 5.25AKK1088 pKa = 10.8INPNTIWYY1096 pKa = 9.69QDD1098 pKa = 3.14ADD1100 pKa = 3.5GDD1102 pKa = 4.49GFGNPSVTIKK1112 pKa = 10.8SCVKK1116 pKa = 7.8PTGYY1120 pKa = 10.57VSNKK1124 pKa = 9.64GDD1126 pKa = 4.36CNDD1129 pKa = 3.78NNPNVHH1135 pKa = 7.02PDD1137 pKa = 3.1ATEE1140 pKa = 3.94VPDD1143 pKa = 6.4GIDD1146 pKa = 3.41NNCNGQIDD1154 pKa = 4.57EE1155 pKa = 4.62SSLVYY1160 pKa = 9.6QVYY1163 pKa = 10.15PNPSVDD1169 pKa = 2.81GRR1171 pKa = 11.84FYY1173 pKa = 11.47LDD1175 pKa = 3.41YY1176 pKa = 11.06QNPADD1181 pKa = 3.8SQIQVVVVSITGTIVYY1197 pKa = 9.03QRR1199 pKa = 11.84NVFLTNGKK1207 pKa = 8.87VQGFFIDD1214 pKa = 3.59MTNQASGSYY1223 pKa = 8.71IVRR1226 pKa = 11.84IISSTTNVSVKK1237 pKa = 10.33VIKK1240 pKa = 10.59LL1241 pKa = 3.43
AA1 pKa = 7.83INPNTQWYY9 pKa = 9.92LDD11 pKa = 3.68NDD13 pKa = 3.51NDD15 pKa = 4.14GFGAGAVAFTQCTKK29 pKa = 9.5PTGNYY34 pKa = 5.87VTNNTDD40 pKa = 3.87CNDD43 pKa = 3.23NDD45 pKa = 3.85TTLNPNTKK53 pKa = 8.86WYY55 pKa = 10.44RR56 pKa = 11.84DD57 pKa = 3.26ADD59 pKa = 3.58GDD61 pKa = 4.59GFGNPADD68 pKa = 4.76EE69 pKa = 4.85KK70 pKa = 9.53TQCLQPAGYY79 pKa = 9.81VRR81 pKa = 11.84NNTDD85 pKa = 3.22CDD87 pKa = 4.24DD88 pKa = 4.15NNVAINPTTVWYY100 pKa = 10.42RR101 pKa = 11.84DD102 pKa = 3.11ADD104 pKa = 3.58GDD106 pKa = 4.44GFGNPAITQVSCTQPTGFVLNNADD130 pKa = 4.77CNDD133 pKa = 3.37SDD135 pKa = 4.38ATLNPNTKK143 pKa = 8.87WYY145 pKa = 10.44RR146 pKa = 11.84DD147 pKa = 3.26ADD149 pKa = 3.58GDD151 pKa = 4.59GFGNPADD158 pKa = 4.76EE159 pKa = 4.85KK160 pKa = 9.53TQCLQPAGYY169 pKa = 9.81VRR171 pKa = 11.84NNTDD175 pKa = 3.43CNDD178 pKa = 3.3NNVAINPTTVWYY190 pKa = 10.39RR191 pKa = 11.84DD192 pKa = 3.17TDD194 pKa = 3.44GDD196 pKa = 4.27GFGNPAITQVSCTQPTGFVLNNADD220 pKa = 4.76CDD222 pKa = 3.94DD223 pKa = 4.08TKK225 pKa = 11.9ADD227 pKa = 3.94INPNTKK233 pKa = 8.89WYY235 pKa = 10.39RR236 pKa = 11.84DD237 pKa = 3.26ADD239 pKa = 3.58GDD241 pKa = 4.59GFGNPADD248 pKa = 4.76EE249 pKa = 4.85KK250 pKa = 9.53TQCLQPAGYY259 pKa = 9.81VRR261 pKa = 11.84NNTDD265 pKa = 3.82CNDD268 pKa = 3.13NNAAINPTTVWYY280 pKa = 10.42RR281 pKa = 11.84DD282 pKa = 3.11ADD284 pKa = 3.58GDD286 pKa = 4.44GFGNPAITQVSCTQPTGFVLNNADD310 pKa = 4.77CNDD313 pKa = 3.37SDD315 pKa = 4.38ATLNPNTKK323 pKa = 8.87WYY325 pKa = 10.44RR326 pKa = 11.84DD327 pKa = 3.26ADD329 pKa = 3.58GDD331 pKa = 4.59GFGNPADD338 pKa = 4.76EE339 pKa = 4.85KK340 pKa = 9.53TQCLQPAGYY349 pKa = 9.81VRR351 pKa = 11.84NNTDD355 pKa = 4.04CDD357 pKa = 3.69DD358 pKa = 3.92TKK360 pKa = 11.9ADD362 pKa = 4.3INPNTQWYY370 pKa = 9.88RR371 pKa = 11.84DD372 pKa = 3.41ADD374 pKa = 3.62GDD376 pKa = 4.59GFGNPADD383 pKa = 4.27VKK385 pKa = 8.09TQCLQPAGYY394 pKa = 9.81VRR396 pKa = 11.84NNTDD400 pKa = 3.43CNDD403 pKa = 3.3NNVAINPTTVWYY415 pKa = 10.42RR416 pKa = 11.84DD417 pKa = 3.11ADD419 pKa = 3.58GDD421 pKa = 4.27GFGNPTITQVSCTQPTGFVLSNADD445 pKa = 4.35CNDD448 pKa = 2.93SDD450 pKa = 4.38ATLNPNTKK458 pKa = 8.87WYY460 pKa = 10.44RR461 pKa = 11.84DD462 pKa = 3.26ADD464 pKa = 3.58GDD466 pKa = 4.59GFGNPADD473 pKa = 5.01AKK475 pKa = 7.57TQCLQPTGYY484 pKa = 10.02IRR486 pKa = 11.84DD487 pKa = 3.8NTDD490 pKa = 3.24CDD492 pKa = 3.7DD493 pKa = 3.87TKK495 pKa = 11.9ADD497 pKa = 4.3INPNTQWYY505 pKa = 9.88RR506 pKa = 11.84DD507 pKa = 3.41ADD509 pKa = 3.62GDD511 pKa = 4.59GFGNPADD518 pKa = 4.26VKK520 pKa = 10.36NQCLQPTGYY529 pKa = 10.47VRR531 pKa = 11.84DD532 pKa = 3.9NTDD535 pKa = 3.72CNDD538 pKa = 3.46SNASLNPNTKK548 pKa = 8.87WYY550 pKa = 10.55ADD552 pKa = 3.51NDD554 pKa = 3.86GDD556 pKa = 4.3GFGAATVAFTQCNKK570 pKa = 9.7PAGNYY575 pKa = 6.2VTNNTDD581 pKa = 4.0CDD583 pKa = 3.8DD584 pKa = 3.92TKK586 pKa = 11.9ADD588 pKa = 3.94INPNTKK594 pKa = 8.89WYY596 pKa = 10.39RR597 pKa = 11.84DD598 pKa = 3.26ADD600 pKa = 3.58GDD602 pKa = 4.74GFGNPANVKK611 pKa = 6.75TQCLQPAGYY620 pKa = 9.95IRR622 pKa = 11.84DD623 pKa = 3.78NTDD626 pKa = 3.55CNDD629 pKa = 3.35SDD631 pKa = 3.85ATLNPNTKK639 pKa = 8.89WYY641 pKa = 10.55ADD643 pKa = 3.51NDD645 pKa = 3.87GDD647 pKa = 4.48GFGAGPVVFVQCAKK661 pKa = 9.28PTGSYY666 pKa = 7.63VTNNGDD672 pKa = 4.62CDD674 pKa = 3.91DD675 pKa = 4.52TNPAIKK681 pKa = 8.61PTAVWYY687 pKa = 10.46RR688 pKa = 11.84DD689 pKa = 3.12ADD691 pKa = 3.56GDD693 pKa = 4.33GFGNPSISVVACSQPAGYY711 pKa = 10.95VMNNTDD717 pKa = 4.82CNDD720 pKa = 3.19NDD722 pKa = 4.34ASINPNTKK730 pKa = 8.92WYY732 pKa = 10.54ADD734 pKa = 3.58NDD736 pKa = 3.62NDD738 pKa = 4.35GFGAGAVAFTQCAKK752 pKa = 10.4PVGNFVTNNSDD763 pKa = 4.61CNDD766 pKa = 3.23NDD768 pKa = 3.38ATLNPNTKK776 pKa = 8.87WYY778 pKa = 10.44RR779 pKa = 11.84DD780 pKa = 3.26ADD782 pKa = 3.58GDD784 pKa = 4.59GFGNPADD791 pKa = 4.27VKK793 pKa = 8.09TQCLQPAGYY802 pKa = 9.95IRR804 pKa = 11.84DD805 pKa = 3.81NTDD808 pKa = 3.24CDD810 pKa = 3.7DD811 pKa = 3.87TKK813 pKa = 11.9ADD815 pKa = 3.94INPNTKK821 pKa = 8.89WYY823 pKa = 10.39RR824 pKa = 11.84DD825 pKa = 3.26ADD827 pKa = 3.58GDD829 pKa = 4.59GFGNPADD836 pKa = 4.06VRR838 pKa = 11.84TQCLQPVGYY847 pKa = 9.96VRR849 pKa = 11.84SNADD853 pKa = 3.9CNDD856 pKa = 3.06SDD858 pKa = 4.38ATLNPNTIWYY868 pKa = 9.54RR869 pKa = 11.84DD870 pKa = 3.26ADD872 pKa = 3.6GDD874 pKa = 4.28GFGNPSVTQKK884 pKa = 10.88SCTQPAGFVRR894 pKa = 11.84NNQDD898 pKa = 3.94CNDD901 pKa = 3.9SDD903 pKa = 5.16ASLNPNTKK911 pKa = 8.84WYY913 pKa = 10.44RR914 pKa = 11.84DD915 pKa = 3.26ADD917 pKa = 3.58GDD919 pKa = 4.59GFGNPADD926 pKa = 4.06VRR928 pKa = 11.84TQCLQPAGYY937 pKa = 9.81VRR939 pKa = 11.84NNTDD943 pKa = 3.72CDD945 pKa = 3.61DD946 pKa = 3.77TNRR949 pKa = 11.84NINPNTKK956 pKa = 9.24WYY958 pKa = 10.43LDD960 pKa = 3.43QDD962 pKa = 3.56GDD964 pKa = 4.86GFGDD968 pKa = 3.58PNNFIVSCTKK978 pKa = 10.27PVGNYY983 pKa = 8.99VLNGTDD989 pKa = 3.42NCPTLPNPNQVIPTWYY1005 pKa = 10.18LDD1007 pKa = 3.31QDD1009 pKa = 3.56RR1010 pKa = 11.84DD1011 pKa = 4.3GLGDD1015 pKa = 3.63PANAIQACSRR1025 pKa = 11.84PDD1027 pKa = 3.43GYY1029 pKa = 11.61VSNNTDD1035 pKa = 3.0NCPTIFNPDD1044 pKa = 2.59GTVPKK1049 pKa = 9.65WYY1051 pKa = 9.87RR1052 pKa = 11.84DD1053 pKa = 3.19ADD1055 pKa = 3.59GDD1057 pKa = 4.39GFGNPNVVVSACTAPIGYY1075 pKa = 9.52VPDD1078 pKa = 3.77NTDD1081 pKa = 4.68CDD1083 pKa = 4.48DD1084 pKa = 4.48SDD1086 pKa = 5.25AKK1088 pKa = 10.8INPNTIWYY1096 pKa = 9.69QDD1098 pKa = 3.14ADD1100 pKa = 3.5GDD1102 pKa = 4.49GFGNPSVTIKK1112 pKa = 10.8SCVKK1116 pKa = 7.8PTGYY1120 pKa = 10.57VSNKK1124 pKa = 9.64GDD1126 pKa = 4.36CNDD1129 pKa = 3.78NNPNVHH1135 pKa = 7.02PDD1137 pKa = 3.1ATEE1140 pKa = 3.94VPDD1143 pKa = 6.4GIDD1146 pKa = 3.41NNCNGQIDD1154 pKa = 4.57EE1155 pKa = 4.62SSLVYY1160 pKa = 9.6QVYY1163 pKa = 10.15PNPSVDD1169 pKa = 2.81GRR1171 pKa = 11.84FYY1173 pKa = 11.47LDD1175 pKa = 3.41YY1176 pKa = 11.06QNPADD1181 pKa = 3.8SQIQVVVVSITGTIVYY1197 pKa = 9.03QRR1199 pKa = 11.84NVFLTNGKK1207 pKa = 8.87VQGFFIDD1214 pKa = 3.59MTNQASGSYY1223 pKa = 8.71IVRR1226 pKa = 11.84IISSTTNVSVKK1237 pKa = 10.33VIKK1240 pKa = 10.59LL1241 pKa = 3.43
Molecular weight: 134.23 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I2HHM9|A0A1I2HHM9_9BACT NADPH:quinone reductase OS=Thermoflexibacter ruber OX=1003 GN=SAMN04488541_102353 PE=4 SV=1
MM1 pKa = 7.59KK2 pKa = 10.21KK3 pKa = 9.99LHH5 pKa = 6.42LLFVKK10 pKa = 10.63ALLLLFLSLVFGTHH24 pKa = 6.42HH25 pKa = 6.21STAQVIIIQPRR36 pKa = 11.84MPQVIVVPPRR46 pKa = 11.84VIITTPRR53 pKa = 11.84GFRR56 pKa = 11.84NPRR59 pKa = 11.84HH60 pKa = 5.03FWGGRR65 pKa = 11.84KK66 pKa = 7.72FQRR69 pKa = 11.84KK70 pKa = 8.86LYY72 pKa = 9.85RR73 pKa = 11.84HH74 pKa = 6.54RR75 pKa = 11.84GWGWRR80 pKa = 11.84RR81 pKa = 11.84WW82 pKa = 3.44
MM1 pKa = 7.59KK2 pKa = 10.21KK3 pKa = 9.99LHH5 pKa = 6.42LLFVKK10 pKa = 10.63ALLLLFLSLVFGTHH24 pKa = 6.42HH25 pKa = 6.21STAQVIIIQPRR36 pKa = 11.84MPQVIVVPPRR46 pKa = 11.84VIITTPRR53 pKa = 11.84GFRR56 pKa = 11.84NPRR59 pKa = 11.84HH60 pKa = 5.03FWGGRR65 pKa = 11.84KK66 pKa = 7.72FQRR69 pKa = 11.84KK70 pKa = 8.86LYY72 pKa = 9.85RR73 pKa = 11.84HH74 pKa = 6.54RR75 pKa = 11.84GWGWRR80 pKa = 11.84RR81 pKa = 11.84WW82 pKa = 3.44
Molecular weight: 9.89 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1625937 |
25 |
5668 |
359.6 |
40.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.773 ± 0.031 | 0.832 ± 0.012 |
4.822 ± 0.025 | 6.644 ± 0.051 |
5.317 ± 0.028 | 6.229 ± 0.043 |
1.776 ± 0.017 | 7.738 ± 0.034 |
7.595 ± 0.055 | 9.667 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.13 ± 0.019 | 5.872 ± 0.049 |
3.54 ± 0.024 | 4.293 ± 0.026 |
3.83 ± 0.025 | 6.051 ± 0.032 |
5.537 ± 0.059 | 6.015 ± 0.031 |
1.169 ± 0.014 | 4.171 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
