
Psychroflexus torquis (strain ATCC 700755 / ACAM 623)
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Psychroflexus; Psychroflexus torquis
Average proteome isoelectric point is 6.54
Get precalculated fractions of proteins
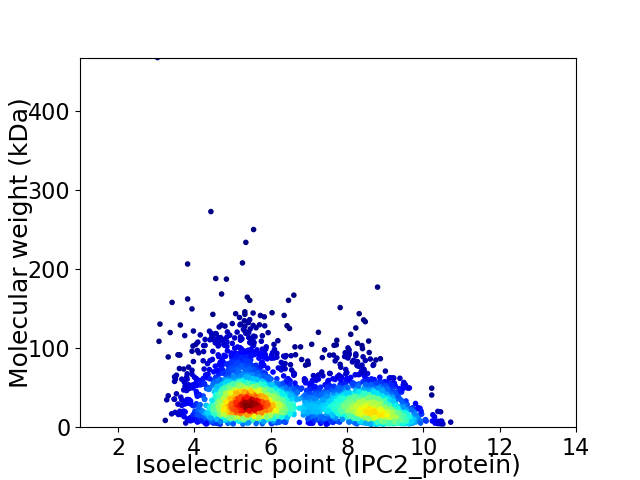
Virtual 2D-PAGE plot for 3457 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
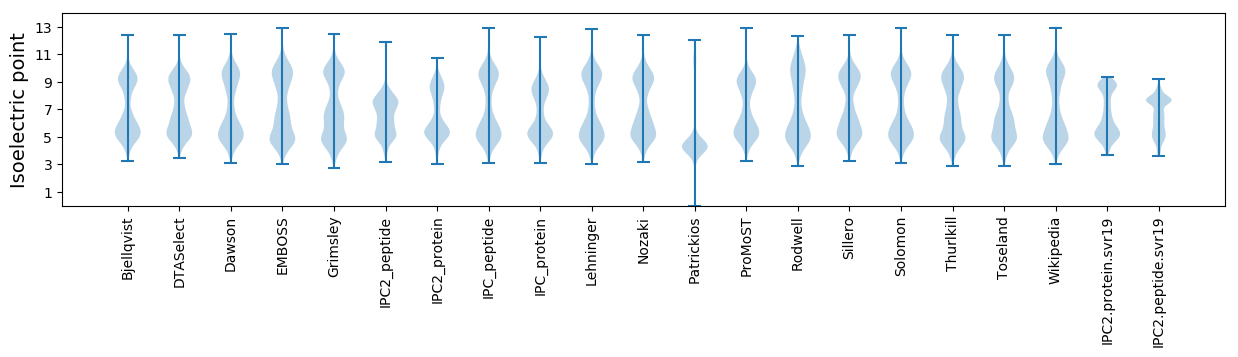
Protein with the lowest isoelectric point:
>tr|K4ID11|K4ID11_PSYTT Uncharacterized protein OS=Psychroflexus torquis (strain ATCC 700755 / ACAM 623) OX=313595 GN=P700755_001382 PE=4 SV=1
MM1 pKa = 7.74KK2 pKa = 10.47NILLLAFTFLFFSSQILSAQSQNKK26 pKa = 9.57HH27 pKa = 5.61IEE29 pKa = 4.19TFLSSKK35 pKa = 10.42VSSKK39 pKa = 10.91KK40 pKa = 10.05LAQNDD45 pKa = 2.74ISEE48 pKa = 4.38YY49 pKa = 10.79VITSQHH55 pKa = 6.27KK56 pKa = 9.78SSISNIEE63 pKa = 3.88HH64 pKa = 6.25IYY66 pKa = 10.17FRR68 pKa = 11.84QSIDD72 pKa = 3.04GVSIVGTEE80 pKa = 4.12SSIHH84 pKa = 5.83VYY86 pKa = 10.22PDD88 pKa = 3.12EE89 pKa = 4.33TLLRR93 pKa = 11.84ANFGFKK99 pKa = 10.36KK100 pKa = 10.54SIADD104 pKa = 3.58KK105 pKa = 11.09VIGAKK110 pKa = 8.24TPKK113 pKa = 10.28LSAEE117 pKa = 3.78LAIRR121 pKa = 11.84RR122 pKa = 11.84VAQQMGYY129 pKa = 8.5YY130 pKa = 9.75QSGGLNSLPKK140 pKa = 9.72STIPNSDD147 pKa = 3.64YY148 pKa = 11.0VFSGAGISEE157 pKa = 4.03VNIPVKK163 pKa = 10.31LVYY166 pKa = 10.61VKK168 pKa = 10.84LEE170 pKa = 3.62QSKK173 pKa = 9.79FVLAWEE179 pKa = 4.47LSIKK183 pKa = 7.19EE184 pKa = 4.52TKK186 pKa = 10.57SSDD189 pKa = 2.76WYY191 pKa = 11.27NFYY194 pKa = 11.52ADD196 pKa = 4.02ASTGKK201 pKa = 10.22IISKK205 pKa = 10.43NNWTVNCLEE214 pKa = 4.17GHH216 pKa = 6.41DD217 pKa = 4.16HH218 pKa = 5.67STHH221 pKa = 6.47NEE223 pKa = 3.9SEE225 pKa = 4.2KK226 pKa = 10.66PKK228 pKa = 10.91AFVSVKK234 pKa = 10.57NKK236 pKa = 7.82PTTTNSGLSQGTYY249 pKa = 10.19NVFPLPVEE257 pKa = 5.12DD258 pKa = 4.67PFSDD262 pKa = 3.42PRR264 pKa = 11.84EE265 pKa = 3.95LVSNSNNALASPFGWHH281 pKa = 6.7DD282 pKa = 3.7TNGSIGAEE290 pKa = 3.85FTVTRR295 pKa = 11.84GNNVNAYY302 pKa = 10.0EE303 pKa = 4.68DD304 pKa = 4.13GNNQGFQPDD313 pKa = 4.09GGSSLSFDD321 pKa = 4.04FPFNPDD327 pKa = 3.1YY328 pKa = 11.59SFGDD332 pKa = 3.53QSEE335 pKa = 4.53SAAITNLFYY344 pKa = 10.68WNNIIHH350 pKa = 7.3DD351 pKa = 4.59IIYY354 pKa = 10.26QYY356 pKa = 11.54GFDD359 pKa = 3.62EE360 pKa = 4.81ASGNFQVNNYY370 pKa = 8.77GNPGVGNDD378 pKa = 3.83PVLAEE383 pKa = 4.31AQDD386 pKa = 3.87GGGTCNANFSRR397 pKa = 11.84APEE400 pKa = 4.76GISPVMQMYY409 pKa = 9.97ICNNRR414 pKa = 11.84DD415 pKa = 2.77GDD417 pKa = 3.79FDD419 pKa = 4.17NGVIIHH425 pKa = 6.98EE426 pKa = 4.4YY427 pKa = 9.58GHH429 pKa = 6.77GISIRR434 pKa = 11.84LSGGSNNSNCLLNDD448 pKa = 3.41EE449 pKa = 4.48QMGEE453 pKa = 4.24GWSDD457 pKa = 2.68WYY459 pKa = 11.28GAMLTIRR466 pKa = 11.84DD467 pKa = 4.3GDD469 pKa = 3.77TGTDD473 pKa = 3.44PRR475 pKa = 11.84PIGNWLFGNDD485 pKa = 3.42EE486 pKa = 4.59DD487 pKa = 5.05GAGIRR492 pKa = 11.84AFPYY496 pKa = 9.69STDD499 pKa = 3.49LAVNPQTYY507 pKa = 11.02DD508 pKa = 3.59DD509 pKa = 4.32IKK511 pKa = 10.29PTFGPHH517 pKa = 6.32PLGSIWATMLWEE529 pKa = 4.29MTWGLIDD536 pKa = 4.68DD537 pKa = 5.03HH538 pKa = 8.32GFDD541 pKa = 3.95PDD543 pKa = 4.44FYY545 pKa = 11.23NGSGGNNVALAIVTEE560 pKa = 4.08ALKK563 pKa = 10.49LQPCSPGFVDD573 pKa = 5.87GRR575 pKa = 11.84DD576 pKa = 3.63AILAADD582 pKa = 3.49QALYY586 pKa = 10.66GGQNFCTIWDD596 pKa = 3.84AFSKK600 pKa = 10.58RR601 pKa = 11.84GLGMSADD608 pKa = 3.51QGSSNNRR615 pKa = 11.84TDD617 pKa = 2.95GTEE620 pKa = 4.21AFDD623 pKa = 4.6SPSTMLTTPEE633 pKa = 4.32TIFCITDD640 pKa = 3.29SNQNLSGGLPLGGVYY655 pKa = 10.25SGSGVTDD662 pKa = 3.18SGDD665 pKa = 3.45GEE667 pKa = 4.83NYY669 pKa = 9.35TFSPLEE675 pKa = 3.97AGIGVHH681 pKa = 6.62TITYY685 pKa = 9.63AAEE688 pKa = 4.33SDD690 pKa = 3.8CSSANQASDD699 pKa = 3.54TIEE702 pKa = 4.18VKK704 pKa = 10.77DD705 pKa = 3.88EE706 pKa = 4.03NPIIEE711 pKa = 4.44CQNISVALDD720 pKa = 3.42EE721 pKa = 6.12DD722 pKa = 4.16GTATIIPQDD731 pKa = 2.91IVTNFEE737 pKa = 3.9ASDD740 pKa = 4.62GYY742 pKa = 9.52TLDD745 pKa = 3.4QSGTFAPEE753 pKa = 4.25NIDD756 pKa = 5.49DD757 pKa = 4.08VDD759 pKa = 4.07TQISLGDD766 pKa = 3.73DD767 pKa = 3.3QVASGLSLGFDD778 pKa = 3.66FNFYY782 pKa = 9.16GTDD785 pKa = 3.36YY786 pKa = 10.9SAFGISSNGYY796 pKa = 10.42LSFSDD801 pKa = 4.6NFDD804 pKa = 3.66SGCCSGQTLPNSNLPNNLIALGWTDD829 pKa = 4.84LNPGNGGSISYY840 pKa = 8.33ATIGSAPNRR849 pKa = 11.84ILIVQFDD856 pKa = 4.21NIQNYY861 pKa = 9.46GDD863 pKa = 3.55SSKK866 pKa = 9.89TVTSQIKK873 pKa = 10.16LFEE876 pKa = 4.55GSNHH880 pKa = 6.19IEE882 pKa = 3.58IHH884 pKa = 5.53SEE886 pKa = 3.82NVEE889 pKa = 4.33GNNMTQGIEE898 pKa = 4.06NTDD901 pKa = 2.77GSMALPVPGRR911 pKa = 11.84NSEE914 pKa = 4.25TLSLSNDD921 pKa = 3.12FVSFIPNTGSFSDD934 pKa = 3.58NCGLEE939 pKa = 4.34TIVTLDD945 pKa = 3.8INSFDD950 pKa = 4.95CNDD953 pKa = 3.53LGEE956 pKa = 4.91NIVTATATDD965 pKa = 3.59TAGNLASCMATVTVTSDD982 pKa = 3.27LDD984 pKa = 3.75VTFSDD989 pKa = 3.86VDD991 pKa = 3.56EE992 pKa = 4.94EE993 pKa = 4.52FCIDD997 pKa = 3.55QNPITGLNGGLPVGGFYY1014 pKa = 10.54SGNGVTDD1021 pKa = 4.25DD1022 pKa = 4.28GNGEE1026 pKa = 4.27TFTFNPSVAGEE1037 pKa = 4.25GTTTITYY1044 pKa = 10.59NGDD1047 pKa = 3.5NSCSAEE1053 pKa = 3.85GSATIDD1059 pKa = 3.59VEE1061 pKa = 4.25IQSAIPVLDD1070 pKa = 4.25CQDD1073 pKa = 2.62ITVPLNIDD1081 pKa = 3.47GLVTITVEE1089 pKa = 4.38DD1090 pKa = 3.77VFGADD1095 pKa = 4.58SIPVDD1100 pKa = 3.55NCVGEE1105 pKa = 4.3PLTFSISPNTFGCDD1119 pKa = 3.02DD1120 pKa = 3.63VGEE1123 pKa = 4.61NIVTVTATDD1132 pKa = 3.39NAGNSQTCTSTVTVEE1147 pKa = 4.21DD1148 pKa = 3.69TAPLQFEE1155 pKa = 5.06CINSIEE1161 pKa = 4.18LVLDD1165 pKa = 3.89EE1166 pKa = 5.0NQTATIKK1173 pKa = 10.75EE1174 pKa = 3.88DD1175 pKa = 4.84DD1176 pKa = 3.93IFEE1179 pKa = 4.65SLPQDD1184 pKa = 3.51SCDD1187 pKa = 3.19NGYY1190 pKa = 10.39SVSISQSNFNCDD1202 pKa = 3.39DD1203 pKa = 4.24LGNSSVEE1210 pKa = 4.06EE1211 pKa = 3.78ILINGSFEE1219 pKa = 4.09NGLTGWTSTVEE1230 pKa = 4.81DD1231 pKa = 4.73GLDD1234 pKa = 3.75DD1235 pKa = 5.33DD1236 pKa = 5.7SPGSCEE1242 pKa = 4.16QAWKK1246 pKa = 9.62PLEE1249 pKa = 4.57DD1250 pKa = 4.12SSTICCCVDD1259 pKa = 3.34NIVPTEE1265 pKa = 4.04GASASFTSFDD1275 pKa = 3.46GQAGTKK1281 pKa = 10.17YY1282 pKa = 10.27ILEE1285 pKa = 4.02QSFIATSGGNAILSFDD1301 pKa = 3.21WVAEE1305 pKa = 4.27FNLNTADD1312 pKa = 3.33IDD1314 pKa = 3.77RR1315 pKa = 11.84SFEE1318 pKa = 3.96VGIFDD1323 pKa = 4.22TNGNLIEE1330 pKa = 4.45TIYY1333 pKa = 10.53TEE1335 pKa = 4.63SITAGEE1341 pKa = 4.32TTSINSSLSFDD1352 pKa = 3.29ISSVLSLLDD1361 pKa = 3.54GEE1363 pKa = 4.6EE1364 pKa = 4.24VVLKK1368 pKa = 8.01FTATIPEE1375 pKa = 4.47TLSGPSKK1382 pKa = 11.25AMLDD1386 pKa = 3.75NVSLIVDD1393 pKa = 4.05SSTIPVEE1400 pKa = 3.94ITVTDD1405 pKa = 3.47AFGNSNTCIVPVTVTDD1421 pKa = 4.18PNEE1424 pKa = 3.87EE1425 pKa = 4.9CILSTDD1431 pKa = 4.26DD1432 pKa = 4.92RR1433 pKa = 11.84FLEE1436 pKa = 4.46NSLSLYY1442 pKa = 9.43PNPADD1447 pKa = 3.41TSFTITWNQDD1457 pKa = 3.0VTVEE1461 pKa = 3.96RR1462 pKa = 11.84LEE1464 pKa = 4.1ILDD1467 pKa = 3.69MTGKK1471 pKa = 10.84LILQRR1476 pKa = 11.84RR1477 pKa = 11.84INISDD1482 pKa = 3.56RR1483 pKa = 11.84QTLVGVSSLSSGVYY1497 pKa = 8.77FVKK1500 pKa = 10.69VSSKK1504 pKa = 10.56NGQSIKK1510 pKa = 10.67KK1511 pKa = 9.81LLVKK1515 pKa = 10.68
MM1 pKa = 7.74KK2 pKa = 10.47NILLLAFTFLFFSSQILSAQSQNKK26 pKa = 9.57HH27 pKa = 5.61IEE29 pKa = 4.19TFLSSKK35 pKa = 10.42VSSKK39 pKa = 10.91KK40 pKa = 10.05LAQNDD45 pKa = 2.74ISEE48 pKa = 4.38YY49 pKa = 10.79VITSQHH55 pKa = 6.27KK56 pKa = 9.78SSISNIEE63 pKa = 3.88HH64 pKa = 6.25IYY66 pKa = 10.17FRR68 pKa = 11.84QSIDD72 pKa = 3.04GVSIVGTEE80 pKa = 4.12SSIHH84 pKa = 5.83VYY86 pKa = 10.22PDD88 pKa = 3.12EE89 pKa = 4.33TLLRR93 pKa = 11.84ANFGFKK99 pKa = 10.36KK100 pKa = 10.54SIADD104 pKa = 3.58KK105 pKa = 11.09VIGAKK110 pKa = 8.24TPKK113 pKa = 10.28LSAEE117 pKa = 3.78LAIRR121 pKa = 11.84RR122 pKa = 11.84VAQQMGYY129 pKa = 8.5YY130 pKa = 9.75QSGGLNSLPKK140 pKa = 9.72STIPNSDD147 pKa = 3.64YY148 pKa = 11.0VFSGAGISEE157 pKa = 4.03VNIPVKK163 pKa = 10.31LVYY166 pKa = 10.61VKK168 pKa = 10.84LEE170 pKa = 3.62QSKK173 pKa = 9.79FVLAWEE179 pKa = 4.47LSIKK183 pKa = 7.19EE184 pKa = 4.52TKK186 pKa = 10.57SSDD189 pKa = 2.76WYY191 pKa = 11.27NFYY194 pKa = 11.52ADD196 pKa = 4.02ASTGKK201 pKa = 10.22IISKK205 pKa = 10.43NNWTVNCLEE214 pKa = 4.17GHH216 pKa = 6.41DD217 pKa = 4.16HH218 pKa = 5.67STHH221 pKa = 6.47NEE223 pKa = 3.9SEE225 pKa = 4.2KK226 pKa = 10.66PKK228 pKa = 10.91AFVSVKK234 pKa = 10.57NKK236 pKa = 7.82PTTTNSGLSQGTYY249 pKa = 10.19NVFPLPVEE257 pKa = 5.12DD258 pKa = 4.67PFSDD262 pKa = 3.42PRR264 pKa = 11.84EE265 pKa = 3.95LVSNSNNALASPFGWHH281 pKa = 6.7DD282 pKa = 3.7TNGSIGAEE290 pKa = 3.85FTVTRR295 pKa = 11.84GNNVNAYY302 pKa = 10.0EE303 pKa = 4.68DD304 pKa = 4.13GNNQGFQPDD313 pKa = 4.09GGSSLSFDD321 pKa = 4.04FPFNPDD327 pKa = 3.1YY328 pKa = 11.59SFGDD332 pKa = 3.53QSEE335 pKa = 4.53SAAITNLFYY344 pKa = 10.68WNNIIHH350 pKa = 7.3DD351 pKa = 4.59IIYY354 pKa = 10.26QYY356 pKa = 11.54GFDD359 pKa = 3.62EE360 pKa = 4.81ASGNFQVNNYY370 pKa = 8.77GNPGVGNDD378 pKa = 3.83PVLAEE383 pKa = 4.31AQDD386 pKa = 3.87GGGTCNANFSRR397 pKa = 11.84APEE400 pKa = 4.76GISPVMQMYY409 pKa = 9.97ICNNRR414 pKa = 11.84DD415 pKa = 2.77GDD417 pKa = 3.79FDD419 pKa = 4.17NGVIIHH425 pKa = 6.98EE426 pKa = 4.4YY427 pKa = 9.58GHH429 pKa = 6.77GISIRR434 pKa = 11.84LSGGSNNSNCLLNDD448 pKa = 3.41EE449 pKa = 4.48QMGEE453 pKa = 4.24GWSDD457 pKa = 2.68WYY459 pKa = 11.28GAMLTIRR466 pKa = 11.84DD467 pKa = 4.3GDD469 pKa = 3.77TGTDD473 pKa = 3.44PRR475 pKa = 11.84PIGNWLFGNDD485 pKa = 3.42EE486 pKa = 4.59DD487 pKa = 5.05GAGIRR492 pKa = 11.84AFPYY496 pKa = 9.69STDD499 pKa = 3.49LAVNPQTYY507 pKa = 11.02DD508 pKa = 3.59DD509 pKa = 4.32IKK511 pKa = 10.29PTFGPHH517 pKa = 6.32PLGSIWATMLWEE529 pKa = 4.29MTWGLIDD536 pKa = 4.68DD537 pKa = 5.03HH538 pKa = 8.32GFDD541 pKa = 3.95PDD543 pKa = 4.44FYY545 pKa = 11.23NGSGGNNVALAIVTEE560 pKa = 4.08ALKK563 pKa = 10.49LQPCSPGFVDD573 pKa = 5.87GRR575 pKa = 11.84DD576 pKa = 3.63AILAADD582 pKa = 3.49QALYY586 pKa = 10.66GGQNFCTIWDD596 pKa = 3.84AFSKK600 pKa = 10.58RR601 pKa = 11.84GLGMSADD608 pKa = 3.51QGSSNNRR615 pKa = 11.84TDD617 pKa = 2.95GTEE620 pKa = 4.21AFDD623 pKa = 4.6SPSTMLTTPEE633 pKa = 4.32TIFCITDD640 pKa = 3.29SNQNLSGGLPLGGVYY655 pKa = 10.25SGSGVTDD662 pKa = 3.18SGDD665 pKa = 3.45GEE667 pKa = 4.83NYY669 pKa = 9.35TFSPLEE675 pKa = 3.97AGIGVHH681 pKa = 6.62TITYY685 pKa = 9.63AAEE688 pKa = 4.33SDD690 pKa = 3.8CSSANQASDD699 pKa = 3.54TIEE702 pKa = 4.18VKK704 pKa = 10.77DD705 pKa = 3.88EE706 pKa = 4.03NPIIEE711 pKa = 4.44CQNISVALDD720 pKa = 3.42EE721 pKa = 6.12DD722 pKa = 4.16GTATIIPQDD731 pKa = 2.91IVTNFEE737 pKa = 3.9ASDD740 pKa = 4.62GYY742 pKa = 9.52TLDD745 pKa = 3.4QSGTFAPEE753 pKa = 4.25NIDD756 pKa = 5.49DD757 pKa = 4.08VDD759 pKa = 4.07TQISLGDD766 pKa = 3.73DD767 pKa = 3.3QVASGLSLGFDD778 pKa = 3.66FNFYY782 pKa = 9.16GTDD785 pKa = 3.36YY786 pKa = 10.9SAFGISSNGYY796 pKa = 10.42LSFSDD801 pKa = 4.6NFDD804 pKa = 3.66SGCCSGQTLPNSNLPNNLIALGWTDD829 pKa = 4.84LNPGNGGSISYY840 pKa = 8.33ATIGSAPNRR849 pKa = 11.84ILIVQFDD856 pKa = 4.21NIQNYY861 pKa = 9.46GDD863 pKa = 3.55SSKK866 pKa = 9.89TVTSQIKK873 pKa = 10.16LFEE876 pKa = 4.55GSNHH880 pKa = 6.19IEE882 pKa = 3.58IHH884 pKa = 5.53SEE886 pKa = 3.82NVEE889 pKa = 4.33GNNMTQGIEE898 pKa = 4.06NTDD901 pKa = 2.77GSMALPVPGRR911 pKa = 11.84NSEE914 pKa = 4.25TLSLSNDD921 pKa = 3.12FVSFIPNTGSFSDD934 pKa = 3.58NCGLEE939 pKa = 4.34TIVTLDD945 pKa = 3.8INSFDD950 pKa = 4.95CNDD953 pKa = 3.53LGEE956 pKa = 4.91NIVTATATDD965 pKa = 3.59TAGNLASCMATVTVTSDD982 pKa = 3.27LDD984 pKa = 3.75VTFSDD989 pKa = 3.86VDD991 pKa = 3.56EE992 pKa = 4.94EE993 pKa = 4.52FCIDD997 pKa = 3.55QNPITGLNGGLPVGGFYY1014 pKa = 10.54SGNGVTDD1021 pKa = 4.25DD1022 pKa = 4.28GNGEE1026 pKa = 4.27TFTFNPSVAGEE1037 pKa = 4.25GTTTITYY1044 pKa = 10.59NGDD1047 pKa = 3.5NSCSAEE1053 pKa = 3.85GSATIDD1059 pKa = 3.59VEE1061 pKa = 4.25IQSAIPVLDD1070 pKa = 4.25CQDD1073 pKa = 2.62ITVPLNIDD1081 pKa = 3.47GLVTITVEE1089 pKa = 4.38DD1090 pKa = 3.77VFGADD1095 pKa = 4.58SIPVDD1100 pKa = 3.55NCVGEE1105 pKa = 4.3PLTFSISPNTFGCDD1119 pKa = 3.02DD1120 pKa = 3.63VGEE1123 pKa = 4.61NIVTVTATDD1132 pKa = 3.39NAGNSQTCTSTVTVEE1147 pKa = 4.21DD1148 pKa = 3.69TAPLQFEE1155 pKa = 5.06CINSIEE1161 pKa = 4.18LVLDD1165 pKa = 3.89EE1166 pKa = 5.0NQTATIKK1173 pKa = 10.75EE1174 pKa = 3.88DD1175 pKa = 4.84DD1176 pKa = 3.93IFEE1179 pKa = 4.65SLPQDD1184 pKa = 3.51SCDD1187 pKa = 3.19NGYY1190 pKa = 10.39SVSISQSNFNCDD1202 pKa = 3.39DD1203 pKa = 4.24LGNSSVEE1210 pKa = 4.06EE1211 pKa = 3.78ILINGSFEE1219 pKa = 4.09NGLTGWTSTVEE1230 pKa = 4.81DD1231 pKa = 4.73GLDD1234 pKa = 3.75DD1235 pKa = 5.33DD1236 pKa = 5.7SPGSCEE1242 pKa = 4.16QAWKK1246 pKa = 9.62PLEE1249 pKa = 4.57DD1250 pKa = 4.12SSTICCCVDD1259 pKa = 3.34NIVPTEE1265 pKa = 4.04GASASFTSFDD1275 pKa = 3.46GQAGTKK1281 pKa = 10.17YY1282 pKa = 10.27ILEE1285 pKa = 4.02QSFIATSGGNAILSFDD1301 pKa = 3.21WVAEE1305 pKa = 4.27FNLNTADD1312 pKa = 3.33IDD1314 pKa = 3.77RR1315 pKa = 11.84SFEE1318 pKa = 3.96VGIFDD1323 pKa = 4.22TNGNLIEE1330 pKa = 4.45TIYY1333 pKa = 10.53TEE1335 pKa = 4.63SITAGEE1341 pKa = 4.32TTSINSSLSFDD1352 pKa = 3.29ISSVLSLLDD1361 pKa = 3.54GEE1363 pKa = 4.6EE1364 pKa = 4.24VVLKK1368 pKa = 8.01FTATIPEE1375 pKa = 4.47TLSGPSKK1382 pKa = 11.25AMLDD1386 pKa = 3.75NVSLIVDD1393 pKa = 4.05SSTIPVEE1400 pKa = 3.94ITVTDD1405 pKa = 3.47AFGNSNTCIVPVTVTDD1421 pKa = 4.18PNEE1424 pKa = 3.87EE1425 pKa = 4.9CILSTDD1431 pKa = 4.26DD1432 pKa = 4.92RR1433 pKa = 11.84FLEE1436 pKa = 4.46NSLSLYY1442 pKa = 9.43PNPADD1447 pKa = 3.41TSFTITWNQDD1457 pKa = 3.0VTVEE1461 pKa = 3.96RR1462 pKa = 11.84LEE1464 pKa = 4.1ILDD1467 pKa = 3.69MTGKK1471 pKa = 10.84LILQRR1476 pKa = 11.84RR1477 pKa = 11.84INISDD1482 pKa = 3.56RR1483 pKa = 11.84QTLVGVSSLSSGVYY1497 pKa = 8.77FVKK1500 pKa = 10.69VSSKK1504 pKa = 10.56NGQSIKK1510 pKa = 10.67KK1511 pKa = 9.81LLVKK1515 pKa = 10.68
Molecular weight: 162.31 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K4IJJ4|K4IJJ4_PSYTT Uncharacterized protein OS=Psychroflexus torquis (strain ATCC 700755 / ACAM 623) OX=313595 GN=P700755_002527 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNKK10 pKa = 8.92KK11 pKa = 9.95RR12 pKa = 11.84KK13 pKa = 8.04NKK15 pKa = 9.86HH16 pKa = 4.03GFRR19 pKa = 11.84EE20 pKa = 4.13RR21 pKa = 11.84MSSVNGRR28 pKa = 11.84KK29 pKa = 8.49VLKK32 pKa = 10.12RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.1GRR39 pKa = 11.84KK40 pKa = 7.97KK41 pKa = 10.65LSVSSEE47 pKa = 3.37MSLRR51 pKa = 11.84KK52 pKa = 9.51RR53 pKa = 3.5
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNKK10 pKa = 8.92KK11 pKa = 9.95RR12 pKa = 11.84KK13 pKa = 8.04NKK15 pKa = 9.86HH16 pKa = 4.03GFRR19 pKa = 11.84EE20 pKa = 4.13RR21 pKa = 11.84MSSVNGRR28 pKa = 11.84KK29 pKa = 8.49VLKK32 pKa = 10.12RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.1GRR39 pKa = 11.84KK40 pKa = 7.97KK41 pKa = 10.65LSVSSEE47 pKa = 3.37MSLRR51 pKa = 11.84KK52 pKa = 9.51RR53 pKa = 3.5
Molecular weight: 6.39 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1142495 |
31 |
4408 |
330.5 |
37.46 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.702 ± 0.042 | 0.688 ± 0.012 |
5.788 ± 0.033 | 6.9 ± 0.037 |
5.517 ± 0.032 | 6.078 ± 0.044 |
1.761 ± 0.023 | 8.089 ± 0.045 |
7.97 ± 0.064 | 9.661 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.127 ± 0.019 | 6.026 ± 0.049 |
3.263 ± 0.023 | 3.543 ± 0.025 |
3.456 ± 0.029 | 7.099 ± 0.036 |
5.52 ± 0.042 | 5.848 ± 0.034 |
1.024 ± 0.014 | 3.939 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
