
Hippeastrum latent virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Tymovirales; Betaflexiviridae; Quinvirinae; Carlavirus
Average proteome isoelectric point is 7.29
Get precalculated fractions of proteins
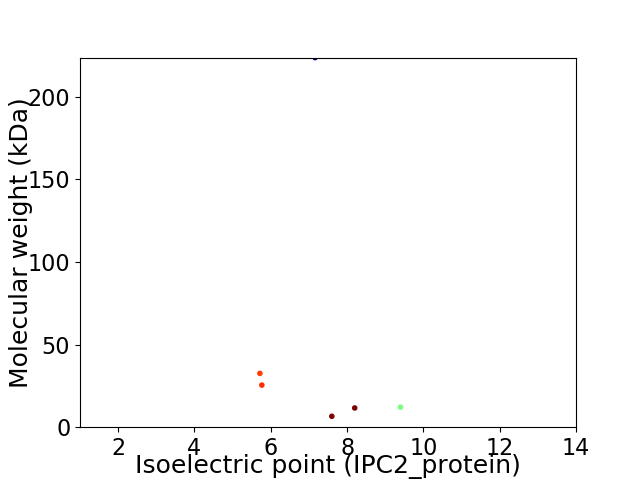
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q4F975|Q4F975_9VIRU 7 kDa protein OS=Hippeastrum latent virus OX=335963 GN=TGB3 PE=3 SV=1
MM1 pKa = 6.62ATKK4 pKa = 10.56VADD7 pKa = 3.8NPSDD11 pKa = 3.99PNVGEE16 pKa = 4.27PKK18 pKa = 9.33ATKK21 pKa = 10.15PGDD24 pKa = 3.89NVDD27 pKa = 3.68PAAAADD33 pKa = 4.13LFDD36 pKa = 5.18DD37 pKa = 4.15ARR39 pKa = 11.84LMEE42 pKa = 4.53DD43 pKa = 3.27QIEE46 pKa = 4.05EE47 pKa = 3.78RR48 pKa = 11.84MARR51 pKa = 11.84LRR53 pKa = 11.84EE54 pKa = 3.98FLMKK58 pKa = 9.54QQRR61 pKa = 11.84AVQVTNPSFEE71 pKa = 4.33LGRR74 pKa = 11.84PKK76 pKa = 10.81LKK78 pKa = 10.2MLDD81 pKa = 3.74SVRR84 pKa = 11.84SDD86 pKa = 3.1PTNLYY91 pKa = 10.11NKK93 pKa = 8.23PTIDD97 pKa = 3.28QLCRR101 pKa = 11.84IRR103 pKa = 11.84PKK105 pKa = 10.68SISNNMATSQDD116 pKa = 3.27MAAITVAIEE125 pKa = 4.06SLGVPSEE132 pKa = 4.15KK133 pKa = 10.48VQTVIIQAVAYY144 pKa = 9.64CKK146 pKa = 10.29DD147 pKa = 3.63ASSSAYY153 pKa = 10.3LDD155 pKa = 3.56PQGTFEE161 pKa = 4.35WEE163 pKa = 4.13GGAIMADD170 pKa = 3.03AVLAILKK177 pKa = 9.88RR178 pKa = 11.84DD179 pKa = 3.26AGTLRR184 pKa = 11.84RR185 pKa = 11.84VCRR188 pKa = 11.84LYY190 pKa = 11.2APVTWNHH197 pKa = 5.31MLAHH201 pKa = 6.71NAPPSDD207 pKa = 3.31WAAMGFQYY215 pKa = 9.65TEE217 pKa = 3.6RR218 pKa = 11.84FAAFDD223 pKa = 3.52CFDD226 pKa = 3.69YY227 pKa = 11.51VEE229 pKa = 4.45NQAAVQPFEE238 pKa = 3.92GLIRR242 pKa = 11.84RR243 pKa = 11.84PTPAEE248 pKa = 4.15KK249 pKa = 9.49IAHH252 pKa = 5.2NTHH255 pKa = 5.92KK256 pKa = 10.71RR257 pKa = 11.84IALDD261 pKa = 3.21RR262 pKa = 11.84ANRR265 pKa = 11.84NEE267 pKa = 4.0RR268 pKa = 11.84FSSLEE273 pKa = 3.68AEE275 pKa = 4.22ITGGRR280 pKa = 11.84LGPEE284 pKa = 3.92IEE286 pKa = 3.77RR287 pKa = 11.84SYY289 pKa = 11.07WKK291 pKa = 9.11TT292 pKa = 3.12
MM1 pKa = 6.62ATKK4 pKa = 10.56VADD7 pKa = 3.8NPSDD11 pKa = 3.99PNVGEE16 pKa = 4.27PKK18 pKa = 9.33ATKK21 pKa = 10.15PGDD24 pKa = 3.89NVDD27 pKa = 3.68PAAAADD33 pKa = 4.13LFDD36 pKa = 5.18DD37 pKa = 4.15ARR39 pKa = 11.84LMEE42 pKa = 4.53DD43 pKa = 3.27QIEE46 pKa = 4.05EE47 pKa = 3.78RR48 pKa = 11.84MARR51 pKa = 11.84LRR53 pKa = 11.84EE54 pKa = 3.98FLMKK58 pKa = 9.54QQRR61 pKa = 11.84AVQVTNPSFEE71 pKa = 4.33LGRR74 pKa = 11.84PKK76 pKa = 10.81LKK78 pKa = 10.2MLDD81 pKa = 3.74SVRR84 pKa = 11.84SDD86 pKa = 3.1PTNLYY91 pKa = 10.11NKK93 pKa = 8.23PTIDD97 pKa = 3.28QLCRR101 pKa = 11.84IRR103 pKa = 11.84PKK105 pKa = 10.68SISNNMATSQDD116 pKa = 3.27MAAITVAIEE125 pKa = 4.06SLGVPSEE132 pKa = 4.15KK133 pKa = 10.48VQTVIIQAVAYY144 pKa = 9.64CKK146 pKa = 10.29DD147 pKa = 3.63ASSSAYY153 pKa = 10.3LDD155 pKa = 3.56PQGTFEE161 pKa = 4.35WEE163 pKa = 4.13GGAIMADD170 pKa = 3.03AVLAILKK177 pKa = 9.88RR178 pKa = 11.84DD179 pKa = 3.26AGTLRR184 pKa = 11.84RR185 pKa = 11.84VCRR188 pKa = 11.84LYY190 pKa = 11.2APVTWNHH197 pKa = 5.31MLAHH201 pKa = 6.71NAPPSDD207 pKa = 3.31WAAMGFQYY215 pKa = 9.65TEE217 pKa = 3.6RR218 pKa = 11.84FAAFDD223 pKa = 3.52CFDD226 pKa = 3.69YY227 pKa = 11.51VEE229 pKa = 4.45NQAAVQPFEE238 pKa = 3.92GLIRR242 pKa = 11.84RR243 pKa = 11.84PTPAEE248 pKa = 4.15KK249 pKa = 9.49IAHH252 pKa = 5.2NTHH255 pKa = 5.92KK256 pKa = 10.71RR257 pKa = 11.84IALDD261 pKa = 3.21RR262 pKa = 11.84ANRR265 pKa = 11.84NEE267 pKa = 4.0RR268 pKa = 11.84FSSLEE273 pKa = 3.68AEE275 pKa = 4.22ITGGRR280 pKa = 11.84LGPEE284 pKa = 3.92IEE286 pKa = 3.77RR287 pKa = 11.84SYY289 pKa = 11.07WKK291 pKa = 9.11TT292 pKa = 3.12
Molecular weight: 32.54 kDa
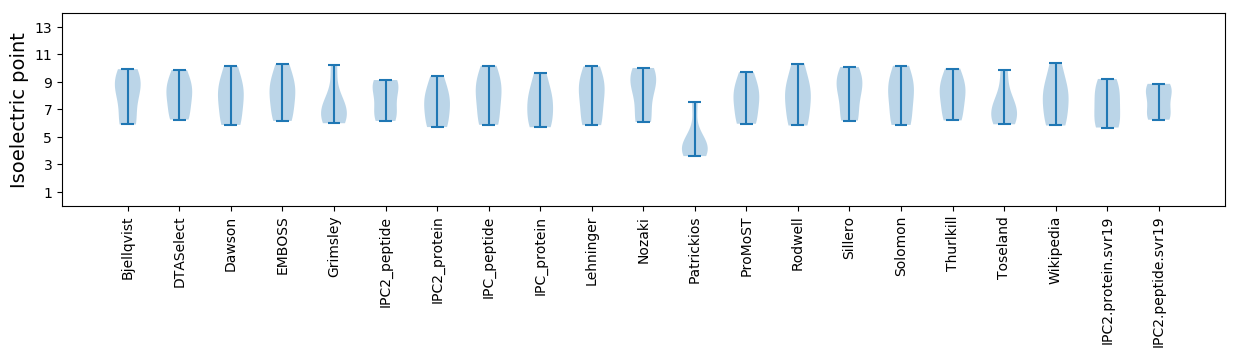
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q4F974|Q4F974_9VIRU Capsid protein OS=Hippeastrum latent virus OX=335963 GN=CP PE=3 SV=1
MM1 pKa = 7.58RR2 pKa = 11.84VDD4 pKa = 4.37ILIALCVNRR13 pKa = 11.84EE14 pKa = 3.74FNKK17 pKa = 10.36RR18 pKa = 11.84GEE20 pKa = 4.09HH21 pKa = 6.45HH22 pKa = 7.18IPLAIYY28 pKa = 8.66IAKK31 pKa = 10.06RR32 pKa = 11.84VGPPLVSTGTSTYY45 pKa = 10.35ARR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84RR50 pKa = 11.84AASIGRR56 pKa = 11.84CHH58 pKa = 6.37RR59 pKa = 11.84CYY61 pKa = 10.06RR62 pKa = 11.84VYY64 pKa = 10.7PPFWWTTRR72 pKa = 11.84CDD74 pKa = 3.51NKK76 pKa = 10.26TCVPGISYY84 pKa = 10.01KK85 pKa = 10.89AEE87 pKa = 3.68VEE89 pKa = 4.85SYY91 pKa = 10.36VKK93 pKa = 10.17WGVAEE98 pKa = 5.45AIPHH102 pKa = 5.98FKK104 pKa = 10.68LL105 pKa = 3.96
MM1 pKa = 7.58RR2 pKa = 11.84VDD4 pKa = 4.37ILIALCVNRR13 pKa = 11.84EE14 pKa = 3.74FNKK17 pKa = 10.36RR18 pKa = 11.84GEE20 pKa = 4.09HH21 pKa = 6.45HH22 pKa = 7.18IPLAIYY28 pKa = 8.66IAKK31 pKa = 10.06RR32 pKa = 11.84VGPPLVSTGTSTYY45 pKa = 10.35ARR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84RR50 pKa = 11.84AASIGRR56 pKa = 11.84CHH58 pKa = 6.37RR59 pKa = 11.84CYY61 pKa = 10.06RR62 pKa = 11.84VYY64 pKa = 10.7PPFWWTTRR72 pKa = 11.84CDD74 pKa = 3.51NKK76 pKa = 10.26TCVPGISYY84 pKa = 10.01KK85 pKa = 10.89AEE87 pKa = 3.68VEE89 pKa = 4.85SYY91 pKa = 10.36VKK93 pKa = 10.17WGVAEE98 pKa = 5.45AIPHH102 pKa = 5.98FKK104 pKa = 10.68LL105 pKa = 3.96
Molecular weight: 12.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2765 |
59 |
1971 |
460.8 |
51.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.21 ± 1.033 | 3.436 ± 0.553 |
4.774 ± 0.632 | 7.052 ± 0.914 |
5.244 ± 0.519 | 6.076 ± 0.423 |
2.351 ± 0.384 | 5.353 ± 0.748 |
5.859 ± 0.762 | 8.969 ± 0.671 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.459 ± 0.493 | 3.508 ± 0.509 |
4.159 ± 1.065 | 3.183 ± 0.377 |
6.799 ± 0.514 | 6.438 ± 0.342 |
4.774 ± 0.621 | 7.052 ± 0.561 |
1.121 ± 0.212 | 3.183 ± 0.363 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
