
Alteribacillus iranensis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillaceae; Alteribacillus
Average proteome isoelectric point is 6.05
Get precalculated fractions of proteins
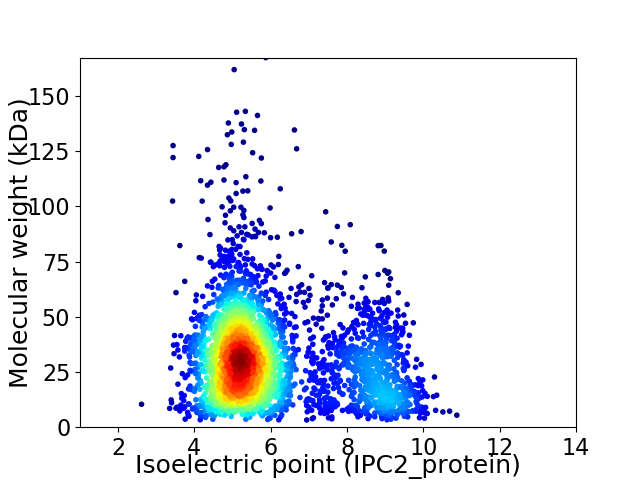
Virtual 2D-PAGE plot for 3218 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
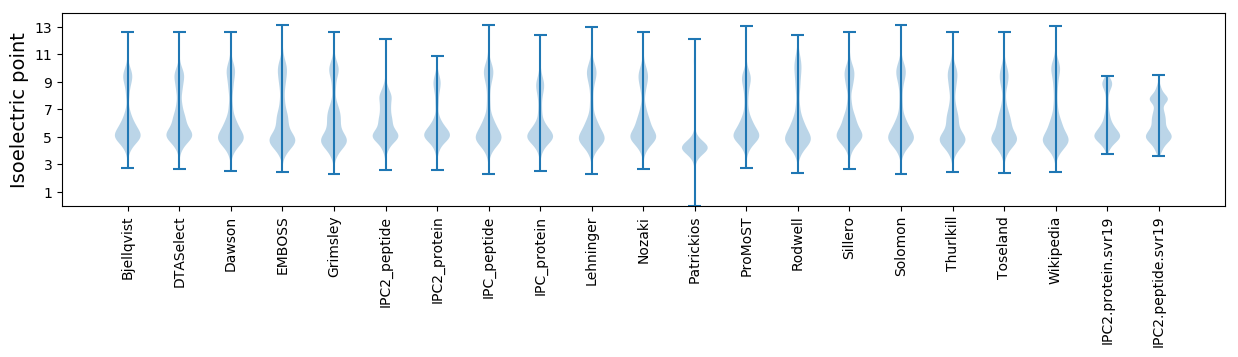
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I2DK74|A0A1I2DK74_9BACI Uncharacterized protein OS=Alteribacillus iranensis OX=930128 GN=SAMN05192532_104113 PE=4 SV=1
MM1 pKa = 7.36KK2 pKa = 10.27RR3 pKa = 11.84LRR5 pKa = 11.84FAALFTASALFIGGCSTSGEE25 pKa = 3.74ADD27 pKa = 3.33QPKK30 pKa = 10.39EE31 pKa = 4.01STEE34 pKa = 4.12GNTQEE39 pKa = 4.39TTTEE43 pKa = 3.87ASSGGGTEE51 pKa = 3.89SGEE54 pKa = 3.97LVIYY58 pKa = 10.7SSGPGGMAEE67 pKa = 3.96EE68 pKa = 4.41LAEE71 pKa = 4.02QFEE74 pKa = 4.59EE75 pKa = 4.22QSGVEE80 pKa = 4.0VEE82 pKa = 5.0LFQSTTGDD90 pKa = 3.24VLGRR94 pKa = 11.84LEE96 pKa = 5.08AEE98 pKa = 4.21QSNPVADD105 pKa = 4.0VVVLASLQPAVDD117 pKa = 3.6YY118 pKa = 10.65MEE120 pKa = 5.29NGLIRR125 pKa = 11.84PYY127 pKa = 10.16EE128 pKa = 4.09SEE130 pKa = 4.41YY131 pKa = 11.33ADD133 pKa = 3.49QLHH136 pKa = 6.87DD137 pKa = 3.18GWYY140 pKa = 10.22DD141 pKa = 3.52EE142 pKa = 6.08DD143 pKa = 4.02YY144 pKa = 11.18HH145 pKa = 8.59FYY147 pKa = 11.06GFSAAALGLSYY158 pKa = 10.2NTDD161 pKa = 4.15LIEE164 pKa = 4.34EE165 pKa = 4.65PPRR168 pKa = 11.84EE169 pKa = 4.35WEE171 pKa = 4.28DD172 pKa = 3.58LTHH175 pKa = 6.91EE176 pKa = 5.08AYY178 pKa = 10.07QDD180 pKa = 3.54QIAIPSPSEE189 pKa = 3.75SGTARR194 pKa = 11.84DD195 pKa = 3.98FVAAYY200 pKa = 9.05IAQEE204 pKa = 4.24GEE206 pKa = 4.01EE207 pKa = 4.16EE208 pKa = 4.19GWAFFEE214 pKa = 4.13EE215 pKa = 4.62LRR217 pKa = 11.84DD218 pKa = 3.7NGLSMEE224 pKa = 4.56GSNNPALTSVINGANGVVMAGVDD247 pKa = 3.42YY248 pKa = 10.41MVYY251 pKa = 10.84NNIEE255 pKa = 3.97GGEE258 pKa = 4.0PLDD261 pKa = 3.58IVFPEE266 pKa = 4.34SGTTITPRR274 pKa = 11.84PAFILEE280 pKa = 3.98SAEE283 pKa = 3.97NVEE286 pKa = 4.24EE287 pKa = 4.07AEE289 pKa = 4.36KK290 pKa = 11.2YY291 pKa = 10.16IDD293 pKa = 5.45FILSDD298 pKa = 3.71EE299 pKa = 4.46GQGIVADD306 pKa = 4.69HH307 pKa = 6.53YY308 pKa = 11.17LLPARR313 pKa = 11.84EE314 pKa = 5.07DD315 pKa = 3.57IPPHH319 pKa = 6.05EE320 pKa = 5.49DD321 pKa = 2.6RR322 pKa = 11.84AAYY325 pKa = 10.12DD326 pKa = 4.1EE327 pKa = 4.16IAEE330 pKa = 5.1LDD332 pKa = 4.08FEE334 pKa = 4.53WEE336 pKa = 4.14YY337 pKa = 11.45LFEE340 pKa = 5.98NGEE343 pKa = 4.64DD344 pKa = 3.19ILSEE348 pKa = 4.12FTEE351 pKa = 4.24MMRR354 pKa = 4.76
MM1 pKa = 7.36KK2 pKa = 10.27RR3 pKa = 11.84LRR5 pKa = 11.84FAALFTASALFIGGCSTSGEE25 pKa = 3.74ADD27 pKa = 3.33QPKK30 pKa = 10.39EE31 pKa = 4.01STEE34 pKa = 4.12GNTQEE39 pKa = 4.39TTTEE43 pKa = 3.87ASSGGGTEE51 pKa = 3.89SGEE54 pKa = 3.97LVIYY58 pKa = 10.7SSGPGGMAEE67 pKa = 3.96EE68 pKa = 4.41LAEE71 pKa = 4.02QFEE74 pKa = 4.59EE75 pKa = 4.22QSGVEE80 pKa = 4.0VEE82 pKa = 5.0LFQSTTGDD90 pKa = 3.24VLGRR94 pKa = 11.84LEE96 pKa = 5.08AEE98 pKa = 4.21QSNPVADD105 pKa = 4.0VVVLASLQPAVDD117 pKa = 3.6YY118 pKa = 10.65MEE120 pKa = 5.29NGLIRR125 pKa = 11.84PYY127 pKa = 10.16EE128 pKa = 4.09SEE130 pKa = 4.41YY131 pKa = 11.33ADD133 pKa = 3.49QLHH136 pKa = 6.87DD137 pKa = 3.18GWYY140 pKa = 10.22DD141 pKa = 3.52EE142 pKa = 6.08DD143 pKa = 4.02YY144 pKa = 11.18HH145 pKa = 8.59FYY147 pKa = 11.06GFSAAALGLSYY158 pKa = 10.2NTDD161 pKa = 4.15LIEE164 pKa = 4.34EE165 pKa = 4.65PPRR168 pKa = 11.84EE169 pKa = 4.35WEE171 pKa = 4.28DD172 pKa = 3.58LTHH175 pKa = 6.91EE176 pKa = 5.08AYY178 pKa = 10.07QDD180 pKa = 3.54QIAIPSPSEE189 pKa = 3.75SGTARR194 pKa = 11.84DD195 pKa = 3.98FVAAYY200 pKa = 9.05IAQEE204 pKa = 4.24GEE206 pKa = 4.01EE207 pKa = 4.16EE208 pKa = 4.19GWAFFEE214 pKa = 4.13EE215 pKa = 4.62LRR217 pKa = 11.84DD218 pKa = 3.7NGLSMEE224 pKa = 4.56GSNNPALTSVINGANGVVMAGVDD247 pKa = 3.42YY248 pKa = 10.41MVYY251 pKa = 10.84NNIEE255 pKa = 3.97GGEE258 pKa = 4.0PLDD261 pKa = 3.58IVFPEE266 pKa = 4.34SGTTITPRR274 pKa = 11.84PAFILEE280 pKa = 3.98SAEE283 pKa = 3.97NVEE286 pKa = 4.24EE287 pKa = 4.07AEE289 pKa = 4.36KK290 pKa = 11.2YY291 pKa = 10.16IDD293 pKa = 5.45FILSDD298 pKa = 3.71EE299 pKa = 4.46GQGIVADD306 pKa = 4.69HH307 pKa = 6.53YY308 pKa = 11.17LLPARR313 pKa = 11.84EE314 pKa = 5.07DD315 pKa = 3.57IPPHH319 pKa = 6.05EE320 pKa = 5.49DD321 pKa = 2.6RR322 pKa = 11.84AAYY325 pKa = 10.12DD326 pKa = 4.1EE327 pKa = 4.16IAEE330 pKa = 5.1LDD332 pKa = 4.08FEE334 pKa = 4.53WEE336 pKa = 4.14YY337 pKa = 11.45LFEE340 pKa = 5.98NGEE343 pKa = 4.64DD344 pKa = 3.19ILSEE348 pKa = 4.12FTEE351 pKa = 4.24MMRR354 pKa = 4.76
Molecular weight: 38.91 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I2C5W2|A0A1I2C5W2_9BACI DNA gyrase subunit B OS=Alteribacillus iranensis OX=930128 GN=gyrB PE=3 SV=1
MM1 pKa = 7.67GKK3 pKa = 7.97PTFKK7 pKa = 10.7PNTRR11 pKa = 11.84KK12 pKa = 9.83RR13 pKa = 11.84KK14 pKa = 8.17KK15 pKa = 8.5VHH17 pKa = 5.6GFRR20 pKa = 11.84EE21 pKa = 3.96RR22 pKa = 11.84MRR24 pKa = 11.84TKK26 pKa = 10.55NGRR29 pKa = 11.84HH30 pKa = 3.86VLKK33 pKa = 10.52NRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 8.66GRR40 pKa = 11.84KK41 pKa = 8.69VLSAA45 pKa = 4.05
MM1 pKa = 7.67GKK3 pKa = 7.97PTFKK7 pKa = 10.7PNTRR11 pKa = 11.84KK12 pKa = 9.83RR13 pKa = 11.84KK14 pKa = 8.17KK15 pKa = 8.5VHH17 pKa = 5.6GFRR20 pKa = 11.84EE21 pKa = 3.96RR22 pKa = 11.84MRR24 pKa = 11.84TKK26 pKa = 10.55NGRR29 pKa = 11.84HH30 pKa = 3.86VLKK33 pKa = 10.52NRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 8.66GRR40 pKa = 11.84KK41 pKa = 8.69VLSAA45 pKa = 4.05
Molecular weight: 5.44 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
934243 |
29 |
1495 |
290.3 |
32.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.29 ± 0.04 | 0.675 ± 0.014 |
5.256 ± 0.041 | 8.273 ± 0.053 |
4.266 ± 0.04 | 7.076 ± 0.037 |
2.292 ± 0.024 | 7.244 ± 0.039 |
6.145 ± 0.048 | 9.428 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.881 ± 0.02 | 3.968 ± 0.027 |
3.854 ± 0.023 | 3.687 ± 0.027 |
4.456 ± 0.03 | 6.099 ± 0.028 |
5.596 ± 0.031 | 7.111 ± 0.035 |
1.077 ± 0.019 | 3.325 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
