
Hubei coleoptera virus 3
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Jingchuvirales; Aliusviridae; Ollusvirus; Ollusvirus coleopteri
Average proteome isoelectric point is 6.2
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
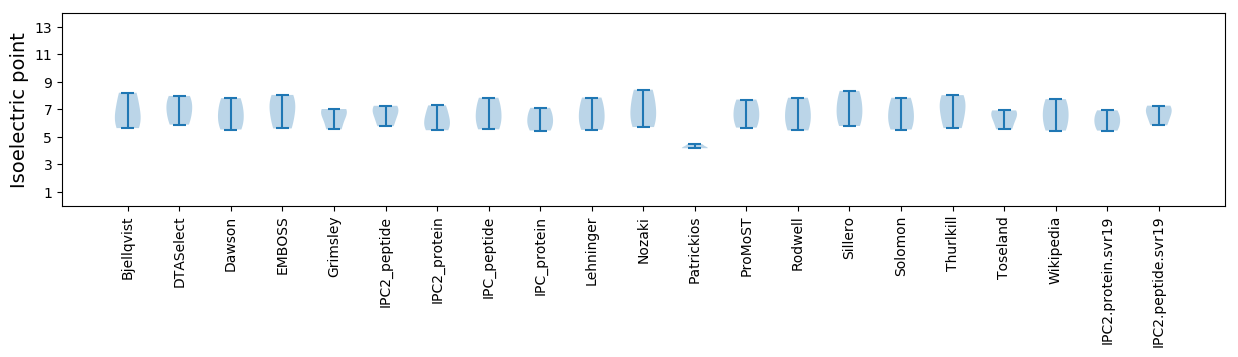
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A1L3KMZ8|A0A1L3KMZ8_9VIRU Putative glycoprotein OS=Hubei coleoptera virus 3 OX=1922862 PE=4 SV=1
MM1 pKa = 7.16FPSRR5 pKa = 11.84QRR7 pKa = 11.84VQLAMNRR14 pKa = 11.84GRR16 pKa = 11.84TIRR19 pKa = 11.84ASPMFAAIGQFSKK32 pKa = 11.02SVDD35 pKa = 2.9IGRR38 pKa = 11.84EE39 pKa = 3.96LLLMKK44 pKa = 10.73AMSRR48 pKa = 11.84INNQPGFVNFSGAILIANQLSKK70 pKa = 10.52IVSSPTGAVTGSTSISQVSGILFARR95 pKa = 11.84WMLTEE100 pKa = 4.54EE101 pKa = 4.17LTEE104 pKa = 4.1NDD106 pKa = 3.13IQSSRR111 pKa = 11.84KK112 pKa = 9.71EE113 pKa = 3.82IVRR116 pKa = 11.84LLDD119 pKa = 3.62LQPPLSGPDD128 pKa = 3.46VPNLLVKK135 pKa = 10.65IMIEE139 pKa = 3.48IGKK142 pKa = 9.21IPKK145 pKa = 10.12DD146 pKa = 3.67SIIDD150 pKa = 3.55HH151 pKa = 7.29DD152 pKa = 4.33GATARR157 pKa = 11.84LGRR160 pKa = 11.84FSCEE164 pKa = 3.15AVIPQEE170 pKa = 3.76GLAARR175 pKa = 11.84KK176 pKa = 9.57VVFRR180 pKa = 11.84EE181 pKa = 3.65YY182 pKa = 10.41RR183 pKa = 11.84GNLIPTTWWGHH194 pKa = 3.97QTNSEE199 pKa = 4.27CANEE203 pKa = 4.26LSTLTTGPTTLDD215 pKa = 3.7FDD217 pKa = 5.24SSLQPMTPDD226 pKa = 3.64TPWASLTDD234 pKa = 4.06DD235 pKa = 3.98DD236 pKa = 5.6AKK238 pKa = 11.25LGMVEE243 pKa = 4.83LYY245 pKa = 10.99GSIHH249 pKa = 5.91SEE251 pKa = 3.63RR252 pKa = 11.84MRR254 pKa = 11.84QIFNSAFVIAVIALSKK270 pKa = 9.69HH271 pKa = 6.83DD272 pKa = 4.33NMTDD276 pKa = 2.67SWLEE280 pKa = 3.78TRR282 pKa = 11.84LEE284 pKa = 3.98RR285 pKa = 11.84LVTCLDD291 pKa = 3.88DD292 pKa = 4.18SQMTTTVTSEE302 pKa = 4.17AIKK305 pKa = 10.65NFYY308 pKa = 10.19RR309 pKa = 11.84RR310 pKa = 11.84YY311 pKa = 8.24QVEE314 pKa = 4.35KK315 pKa = 10.62IKK317 pKa = 11.12VDD319 pKa = 3.59DD320 pKa = 4.81LYY322 pKa = 11.75DD323 pKa = 3.75LFVYY327 pKa = 10.41INSLCLDD334 pKa = 4.16FDD336 pKa = 4.86LEE338 pKa = 4.21PMQWILEE345 pKa = 4.09QAFVNNVASATAIAEE360 pKa = 4.31IIAKK364 pKa = 10.04SKK366 pKa = 8.95TVPITTILEE375 pKa = 4.26TLPATQIQSLITLVAHH391 pKa = 6.56LQHH394 pKa = 7.3DD395 pKa = 4.66PFCAIIRR402 pKa = 11.84PPITVRR408 pKa = 11.84QYY410 pKa = 11.2PDD412 pKa = 2.77VAYY415 pKa = 10.77VGIAATYY422 pKa = 10.03NSRR425 pKa = 11.84TGPGGKK431 pKa = 9.08RR432 pKa = 11.84YY433 pKa = 9.43QGKK436 pKa = 8.63PDD438 pKa = 3.28TMATLTTAALNGLVNFLLQTSRR460 pKa = 11.84TQMEE464 pKa = 4.27MEE466 pKa = 4.94LNLDD470 pKa = 3.99TYY472 pKa = 11.32AKK474 pKa = 9.72KK475 pKa = 10.45FLMGQDD481 pKa = 3.08NSQVVFIDD489 pKa = 2.95QDD491 pKa = 4.19YY492 pKa = 10.76YY493 pKa = 11.12VIKK496 pKa = 9.95PDD498 pKa = 3.85VVQQPTAGQSGTSTTPTQTDD518 pKa = 3.65VEE520 pKa = 4.28ISRR523 pKa = 11.84DD524 pKa = 3.68LKK526 pKa = 11.15DD527 pKa = 3.15QLLRR531 pKa = 11.84SEE533 pKa = 4.18RR534 pKa = 11.84ADD536 pKa = 2.98WPRR539 pKa = 11.84RR540 pKa = 11.84AKK542 pKa = 10.25SLPEE546 pKa = 3.53GTVRR550 pKa = 11.84MTQQEE555 pKa = 4.42VFAQIVDD562 pKa = 3.84KK563 pKa = 11.21EE564 pKa = 4.29PVKK567 pKa = 10.64MKK569 pKa = 10.67AFRR572 pKa = 11.84DD573 pKa = 3.7AMDD576 pKa = 3.64ILLDD580 pKa = 3.4ITQQKK585 pKa = 9.09PISVLTGTNATARR598 pKa = 11.84KK599 pKa = 8.61RR600 pKa = 11.84KK601 pKa = 9.57SKK603 pKa = 10.04ISGRR607 pKa = 11.84MRR609 pKa = 11.84EE610 pKa = 4.52IIVSWGVTMEE620 pKa = 4.72EE621 pKa = 3.47IWNTEE626 pKa = 4.0DD627 pKa = 3.97APADD631 pKa = 3.94LTHH634 pKa = 5.96GTIDD638 pKa = 3.43IDD640 pKa = 3.84EE641 pKa = 4.43LVCLGRR647 pKa = 11.84KK648 pKa = 9.23KK649 pKa = 10.43IGVAGAQGPAQAPGPAEE666 pKa = 3.91VVEE669 pKa = 4.22QANAA673 pKa = 3.38
MM1 pKa = 7.16FPSRR5 pKa = 11.84QRR7 pKa = 11.84VQLAMNRR14 pKa = 11.84GRR16 pKa = 11.84TIRR19 pKa = 11.84ASPMFAAIGQFSKK32 pKa = 11.02SVDD35 pKa = 2.9IGRR38 pKa = 11.84EE39 pKa = 3.96LLLMKK44 pKa = 10.73AMSRR48 pKa = 11.84INNQPGFVNFSGAILIANQLSKK70 pKa = 10.52IVSSPTGAVTGSTSISQVSGILFARR95 pKa = 11.84WMLTEE100 pKa = 4.54EE101 pKa = 4.17LTEE104 pKa = 4.1NDD106 pKa = 3.13IQSSRR111 pKa = 11.84KK112 pKa = 9.71EE113 pKa = 3.82IVRR116 pKa = 11.84LLDD119 pKa = 3.62LQPPLSGPDD128 pKa = 3.46VPNLLVKK135 pKa = 10.65IMIEE139 pKa = 3.48IGKK142 pKa = 9.21IPKK145 pKa = 10.12DD146 pKa = 3.67SIIDD150 pKa = 3.55HH151 pKa = 7.29DD152 pKa = 4.33GATARR157 pKa = 11.84LGRR160 pKa = 11.84FSCEE164 pKa = 3.15AVIPQEE170 pKa = 3.76GLAARR175 pKa = 11.84KK176 pKa = 9.57VVFRR180 pKa = 11.84EE181 pKa = 3.65YY182 pKa = 10.41RR183 pKa = 11.84GNLIPTTWWGHH194 pKa = 3.97QTNSEE199 pKa = 4.27CANEE203 pKa = 4.26LSTLTTGPTTLDD215 pKa = 3.7FDD217 pKa = 5.24SSLQPMTPDD226 pKa = 3.64TPWASLTDD234 pKa = 4.06DD235 pKa = 3.98DD236 pKa = 5.6AKK238 pKa = 11.25LGMVEE243 pKa = 4.83LYY245 pKa = 10.99GSIHH249 pKa = 5.91SEE251 pKa = 3.63RR252 pKa = 11.84MRR254 pKa = 11.84QIFNSAFVIAVIALSKK270 pKa = 9.69HH271 pKa = 6.83DD272 pKa = 4.33NMTDD276 pKa = 2.67SWLEE280 pKa = 3.78TRR282 pKa = 11.84LEE284 pKa = 3.98RR285 pKa = 11.84LVTCLDD291 pKa = 3.88DD292 pKa = 4.18SQMTTTVTSEE302 pKa = 4.17AIKK305 pKa = 10.65NFYY308 pKa = 10.19RR309 pKa = 11.84RR310 pKa = 11.84YY311 pKa = 8.24QVEE314 pKa = 4.35KK315 pKa = 10.62IKK317 pKa = 11.12VDD319 pKa = 3.59DD320 pKa = 4.81LYY322 pKa = 11.75DD323 pKa = 3.75LFVYY327 pKa = 10.41INSLCLDD334 pKa = 4.16FDD336 pKa = 4.86LEE338 pKa = 4.21PMQWILEE345 pKa = 4.09QAFVNNVASATAIAEE360 pKa = 4.31IIAKK364 pKa = 10.04SKK366 pKa = 8.95TVPITTILEE375 pKa = 4.26TLPATQIQSLITLVAHH391 pKa = 6.56LQHH394 pKa = 7.3DD395 pKa = 4.66PFCAIIRR402 pKa = 11.84PPITVRR408 pKa = 11.84QYY410 pKa = 11.2PDD412 pKa = 2.77VAYY415 pKa = 10.77VGIAATYY422 pKa = 10.03NSRR425 pKa = 11.84TGPGGKK431 pKa = 9.08RR432 pKa = 11.84YY433 pKa = 9.43QGKK436 pKa = 8.63PDD438 pKa = 3.28TMATLTTAALNGLVNFLLQTSRR460 pKa = 11.84TQMEE464 pKa = 4.27MEE466 pKa = 4.94LNLDD470 pKa = 3.99TYY472 pKa = 11.32AKK474 pKa = 9.72KK475 pKa = 10.45FLMGQDD481 pKa = 3.08NSQVVFIDD489 pKa = 2.95QDD491 pKa = 4.19YY492 pKa = 10.76YY493 pKa = 11.12VIKK496 pKa = 9.95PDD498 pKa = 3.85VVQQPTAGQSGTSTTPTQTDD518 pKa = 3.65VEE520 pKa = 4.28ISRR523 pKa = 11.84DD524 pKa = 3.68LKK526 pKa = 11.15DD527 pKa = 3.15QLLRR531 pKa = 11.84SEE533 pKa = 4.18RR534 pKa = 11.84ADD536 pKa = 2.98WPRR539 pKa = 11.84RR540 pKa = 11.84AKK542 pKa = 10.25SLPEE546 pKa = 3.53GTVRR550 pKa = 11.84MTQQEE555 pKa = 4.42VFAQIVDD562 pKa = 3.84KK563 pKa = 11.21EE564 pKa = 4.29PVKK567 pKa = 10.64MKK569 pKa = 10.67AFRR572 pKa = 11.84DD573 pKa = 3.7AMDD576 pKa = 3.64ILLDD580 pKa = 3.4ITQQKK585 pKa = 9.09PISVLTGTNATARR598 pKa = 11.84KK599 pKa = 8.61RR600 pKa = 11.84KK601 pKa = 9.57SKK603 pKa = 10.04ISGRR607 pKa = 11.84MRR609 pKa = 11.84EE610 pKa = 4.52IIVSWGVTMEE620 pKa = 4.72EE621 pKa = 3.47IWNTEE626 pKa = 4.0DD627 pKa = 3.97APADD631 pKa = 3.94LTHH634 pKa = 5.96GTIDD638 pKa = 3.43IDD640 pKa = 3.84EE641 pKa = 4.43LVCLGRR647 pKa = 11.84KK648 pKa = 9.23KK649 pKa = 10.43IGVAGAQGPAQAPGPAEE666 pKa = 3.91VVEE669 pKa = 4.22QANAA673 pKa = 3.38
Molecular weight: 74.61 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KMZ8|A0A1L3KMZ8_9VIRU Putative glycoprotein OS=Hubei coleoptera virus 3 OX=1922862 PE=4 SV=1
MM1 pKa = 7.32FFAILLAILTLSYY14 pKa = 11.02GSPTGRR20 pKa = 11.84SKK22 pKa = 11.59LNLEE26 pKa = 4.33TATVMTQHH34 pKa = 7.27DD35 pKa = 4.94PMFLPYY41 pKa = 10.15FEE43 pKa = 4.89WKK45 pKa = 10.18HH46 pKa = 5.4LLHH49 pKa = 6.38VQLLCPDD56 pKa = 4.03SNIMSLIDD64 pKa = 3.82ALTKK68 pKa = 10.58NVYY71 pKa = 10.26DD72 pKa = 3.55QTNKK76 pKa = 10.72LEE78 pKa = 4.78LNLQVLTEE86 pKa = 4.13EE87 pKa = 5.22LSKK90 pKa = 11.6DD91 pKa = 3.51MNNNTLSTNTLLMRR105 pKa = 11.84DD106 pKa = 3.63QVHH109 pKa = 5.5VAKK112 pKa = 10.64NLQAFLRR119 pKa = 11.84GTSEE123 pKa = 3.5KK124 pKa = 9.59WKK126 pKa = 10.57KK127 pKa = 9.95LVNGLFKK134 pKa = 10.56FHH136 pKa = 6.64GMAQQSTCYY145 pKa = 10.26HH146 pKa = 5.98LVKK149 pKa = 9.93TNQGKK154 pKa = 7.1TVPTGISGMASDD166 pKa = 5.25ASTSYY171 pKa = 10.92EE172 pKa = 3.73GDD174 pKa = 3.04LWKK177 pKa = 10.75VLHH180 pKa = 7.5DD181 pKa = 4.05SMTKK185 pKa = 9.78SRR187 pKa = 11.84QKK189 pKa = 9.83RR190 pKa = 11.84TIHH193 pKa = 5.67IHH195 pKa = 6.25GSDD198 pKa = 2.8NTIFHH203 pKa = 7.06NSQITKK209 pKa = 9.93PSDD212 pKa = 2.86ITKK215 pKa = 10.05EE216 pKa = 3.81LRR218 pKa = 11.84RR219 pKa = 11.84DD220 pKa = 3.27KK221 pKa = 11.0DD222 pKa = 3.56LSDD225 pKa = 3.71NPTTVKK231 pKa = 10.34SVSYY235 pKa = 9.95HH236 pKa = 5.21NPKK239 pKa = 10.0SSEE242 pKa = 3.86MLKK245 pKa = 10.51LQNCFCWDD253 pKa = 3.33EE254 pKa = 4.21ATQQAHH260 pKa = 5.6VDD262 pKa = 4.06RR263 pKa = 11.84NCDD266 pKa = 3.2LANCKK271 pKa = 9.68RR272 pKa = 11.84VLDD275 pKa = 3.63KK276 pKa = 10.92WLMKK280 pKa = 10.8GFGQITGNVKK290 pKa = 10.5LYY292 pKa = 10.7NEE294 pKa = 4.14YY295 pKa = 10.66GKK297 pKa = 8.21TVIKK301 pKa = 8.19TTTQPPSQVEE311 pKa = 4.28DD312 pKa = 3.82EE313 pKa = 4.81VVGSPDD319 pKa = 2.93QSEE322 pKa = 4.51VSRR325 pKa = 11.84EE326 pKa = 3.61VDD328 pKa = 3.4GQAEE332 pKa = 4.13TLAEE336 pKa = 4.04KK337 pKa = 10.62LLRR340 pKa = 11.84KK341 pKa = 9.94HH342 pKa = 6.0PVVPSKK348 pKa = 9.99TMADD352 pKa = 3.29ANQASRR358 pKa = 11.84NLPTVDD364 pKa = 3.34EE365 pKa = 4.34QYY367 pKa = 11.37EE368 pKa = 4.21QILLEE373 pKa = 4.31NYY375 pKa = 6.68QTSAFVYY382 pKa = 7.35QLCRR386 pKa = 11.84LRR388 pKa = 11.84LSDD391 pKa = 4.8NLQNCLGTIPEE402 pKa = 4.34TNALNHH408 pKa = 5.42LWEE411 pKa = 4.96RR412 pKa = 11.84LQSTHH417 pKa = 5.58SKK419 pKa = 10.04RR420 pKa = 11.84IKK422 pKa = 9.86RR423 pKa = 11.84SSNRR427 pKa = 11.84VKK429 pKa = 10.55RR430 pKa = 11.84APLGIIGRR438 pKa = 11.84LYY440 pKa = 10.13KK441 pKa = 10.06WLFGVATVDD450 pKa = 4.24DD451 pKa = 4.03VDD453 pKa = 3.56EE454 pKa = 4.14VRR456 pKa = 11.84RR457 pKa = 11.84NFVSYY462 pKa = 11.1LKK464 pKa = 10.62GQQNLSKK471 pKa = 10.81ANQEE475 pKa = 3.91FMTNQLSINQHH486 pKa = 5.13FDD488 pKa = 3.35EE489 pKa = 4.73NLKK492 pKa = 10.32ILNTKK497 pKa = 10.05ISDD500 pKa = 3.79TVKK503 pKa = 10.8GMNSTIDD510 pKa = 3.37MFMDD514 pKa = 5.39TIQQTNNVFQDD525 pKa = 3.97LYY527 pKa = 10.39LTTKK531 pKa = 10.61LSSLLIHH538 pKa = 6.4GNILITGAVIDD549 pKa = 4.21ALRR552 pKa = 11.84INQYY556 pKa = 9.37MDD558 pKa = 3.68SLSSRR563 pKa = 11.84YY564 pKa = 9.71LSLYY568 pKa = 9.86HH569 pKa = 7.18ALLSDD574 pKa = 3.71TLTAEE579 pKa = 4.14ILDD582 pKa = 4.14PASLKK587 pKa = 10.24TILLEE592 pKa = 4.52INNKK596 pKa = 9.62LPPNYY601 pKa = 9.77QVIDD605 pKa = 4.03LPDD608 pKa = 3.44FRR610 pKa = 11.84SWYY613 pKa = 8.42KK614 pKa = 10.35VSYY617 pKa = 9.07STVSLIDD624 pKa = 3.29NSIVYY629 pKa = 9.89AFKK632 pKa = 10.69FPIVRR637 pKa = 11.84KK638 pKa = 10.0SSEE641 pKa = 3.61PTLWHH646 pKa = 6.44FNSLPVFKK654 pKa = 10.86NGTAFKK660 pKa = 10.74LNYY663 pKa = 8.43HH664 pKa = 6.01DD665 pKa = 3.37HH666 pKa = 6.03WVYY669 pKa = 11.14LDD671 pKa = 3.38EE672 pKa = 6.27HH673 pKa = 5.37NQEE676 pKa = 4.33WVPMSQTDD684 pKa = 3.56FNARR688 pKa = 11.84CDD690 pKa = 3.59KK691 pKa = 10.14STSVCSGFASRR702 pKa = 11.84YY703 pKa = 7.2TYY705 pKa = 11.19SKK707 pKa = 11.31NNYY710 pKa = 6.96NHH712 pKa = 6.99CLLSLLYY719 pKa = 10.45DD720 pKa = 3.49SSKK723 pKa = 11.37VIDD726 pKa = 3.85QCEE729 pKa = 4.01MSPIDD734 pKa = 3.78IDD736 pKa = 3.93QVLDD740 pKa = 4.25PIIAISIPPNQWLVEE755 pKa = 3.84TLYY758 pKa = 10.28PVKK761 pKa = 10.36AVKK764 pKa = 10.16SCPAKK769 pKa = 10.3SGQFITDD776 pKa = 3.01SVMISGVMTIEE787 pKa = 4.58LAPLCKK793 pKa = 9.75ISIAEE798 pKa = 4.11LTLEE802 pKa = 4.61SSLAPPGLEE811 pKa = 4.0SQSYY815 pKa = 8.62APEE818 pKa = 3.94YY819 pKa = 9.39TLEE822 pKa = 4.14SVKK825 pKa = 10.52ISTSDD830 pKa = 2.85IDD832 pKa = 4.65IIALKK837 pKa = 9.56EE838 pKa = 4.11NKK840 pKa = 9.37VPQIDD845 pKa = 3.23IKK847 pKa = 10.38MDD849 pKa = 2.91KK850 pKa = 9.7STWNTKK856 pKa = 9.44AMQIKK861 pKa = 9.67IKK863 pKa = 10.72SVEE866 pKa = 4.27TNLNAQKK873 pKa = 10.54EE874 pKa = 4.41LVSNISEE881 pKa = 4.2INKK884 pKa = 9.59KK885 pKa = 9.89LQADD889 pKa = 4.76LIKK892 pKa = 10.34IEE894 pKa = 4.23SSSGKK899 pKa = 8.16TFSSQWISFYY909 pKa = 10.1WEE911 pKa = 3.91NIIFYY916 pKa = 9.15IWVIILTLMTLRR928 pKa = 11.84VYY930 pKa = 10.85VRR932 pKa = 11.84VFGIPLIGRR941 pKa = 11.84GNGIMAKK948 pKa = 8.96ATFPLVSSSGFGTANAFPPQGKK970 pKa = 9.22IYY972 pKa = 9.82MDD974 pKa = 4.74GSTIWASAPNATSNVMVNVTNKK996 pKa = 9.61FNEE999 pKa = 4.71FFSQPHH1005 pKa = 5.28NTTSVIHH1012 pKa = 5.97LCVMVALMLLMLILMYY1028 pKa = 11.19SLMCHH1033 pKa = 5.64HH1034 pKa = 6.91QSMLLRR1040 pKa = 11.84KK1041 pKa = 9.13LLSLGVLPINTRR1053 pKa = 11.84EE1054 pKa = 4.02SQHH1057 pKa = 7.4IGEE1060 pKa = 5.17IGVQLYY1066 pKa = 9.86VLVMMKK1072 pKa = 10.79SMFGSKK1078 pKa = 7.15THH1080 pKa = 5.45YY1081 pKa = 10.85AEE1083 pKa = 5.56VGIQMCTLPGNIRR1096 pKa = 11.84DD1097 pKa = 4.45WICTSNLSEE1106 pKa = 4.05NKK1108 pKa = 9.33TFIVPKK1114 pKa = 9.41WVCRR1118 pKa = 11.84WSSTLEE1124 pKa = 4.41LSCQWGPICLKK1135 pKa = 8.65STKK1138 pKa = 9.91YY1139 pKa = 10.71PNVDD1143 pKa = 3.08TCKK1146 pKa = 10.43DD1147 pKa = 3.72LPKK1150 pKa = 10.46HH1151 pKa = 5.97GSFNMRR1157 pKa = 11.84EE1158 pKa = 4.28VIEE1161 pKa = 4.33QINPKK1166 pKa = 10.34CPFIWKK1172 pKa = 9.44YY1173 pKa = 10.12ISAVEE1178 pKa = 4.05VNKK1181 pKa = 10.43VILRR1185 pKa = 11.84THH1187 pKa = 6.43NDD1189 pKa = 2.84YY1190 pKa = 11.47KK1191 pKa = 10.87EE1192 pKa = 4.1IYY1194 pKa = 9.53SYY1196 pKa = 11.79NSDD1199 pKa = 3.64TLSS1202 pKa = 3.13
MM1 pKa = 7.32FFAILLAILTLSYY14 pKa = 11.02GSPTGRR20 pKa = 11.84SKK22 pKa = 11.59LNLEE26 pKa = 4.33TATVMTQHH34 pKa = 7.27DD35 pKa = 4.94PMFLPYY41 pKa = 10.15FEE43 pKa = 4.89WKK45 pKa = 10.18HH46 pKa = 5.4LLHH49 pKa = 6.38VQLLCPDD56 pKa = 4.03SNIMSLIDD64 pKa = 3.82ALTKK68 pKa = 10.58NVYY71 pKa = 10.26DD72 pKa = 3.55QTNKK76 pKa = 10.72LEE78 pKa = 4.78LNLQVLTEE86 pKa = 4.13EE87 pKa = 5.22LSKK90 pKa = 11.6DD91 pKa = 3.51MNNNTLSTNTLLMRR105 pKa = 11.84DD106 pKa = 3.63QVHH109 pKa = 5.5VAKK112 pKa = 10.64NLQAFLRR119 pKa = 11.84GTSEE123 pKa = 3.5KK124 pKa = 9.59WKK126 pKa = 10.57KK127 pKa = 9.95LVNGLFKK134 pKa = 10.56FHH136 pKa = 6.64GMAQQSTCYY145 pKa = 10.26HH146 pKa = 5.98LVKK149 pKa = 9.93TNQGKK154 pKa = 7.1TVPTGISGMASDD166 pKa = 5.25ASTSYY171 pKa = 10.92EE172 pKa = 3.73GDD174 pKa = 3.04LWKK177 pKa = 10.75VLHH180 pKa = 7.5DD181 pKa = 4.05SMTKK185 pKa = 9.78SRR187 pKa = 11.84QKK189 pKa = 9.83RR190 pKa = 11.84TIHH193 pKa = 5.67IHH195 pKa = 6.25GSDD198 pKa = 2.8NTIFHH203 pKa = 7.06NSQITKK209 pKa = 9.93PSDD212 pKa = 2.86ITKK215 pKa = 10.05EE216 pKa = 3.81LRR218 pKa = 11.84RR219 pKa = 11.84DD220 pKa = 3.27KK221 pKa = 11.0DD222 pKa = 3.56LSDD225 pKa = 3.71NPTTVKK231 pKa = 10.34SVSYY235 pKa = 9.95HH236 pKa = 5.21NPKK239 pKa = 10.0SSEE242 pKa = 3.86MLKK245 pKa = 10.51LQNCFCWDD253 pKa = 3.33EE254 pKa = 4.21ATQQAHH260 pKa = 5.6VDD262 pKa = 4.06RR263 pKa = 11.84NCDD266 pKa = 3.2LANCKK271 pKa = 9.68RR272 pKa = 11.84VLDD275 pKa = 3.63KK276 pKa = 10.92WLMKK280 pKa = 10.8GFGQITGNVKK290 pKa = 10.5LYY292 pKa = 10.7NEE294 pKa = 4.14YY295 pKa = 10.66GKK297 pKa = 8.21TVIKK301 pKa = 8.19TTTQPPSQVEE311 pKa = 4.28DD312 pKa = 3.82EE313 pKa = 4.81VVGSPDD319 pKa = 2.93QSEE322 pKa = 4.51VSRR325 pKa = 11.84EE326 pKa = 3.61VDD328 pKa = 3.4GQAEE332 pKa = 4.13TLAEE336 pKa = 4.04KK337 pKa = 10.62LLRR340 pKa = 11.84KK341 pKa = 9.94HH342 pKa = 6.0PVVPSKK348 pKa = 9.99TMADD352 pKa = 3.29ANQASRR358 pKa = 11.84NLPTVDD364 pKa = 3.34EE365 pKa = 4.34QYY367 pKa = 11.37EE368 pKa = 4.21QILLEE373 pKa = 4.31NYY375 pKa = 6.68QTSAFVYY382 pKa = 7.35QLCRR386 pKa = 11.84LRR388 pKa = 11.84LSDD391 pKa = 4.8NLQNCLGTIPEE402 pKa = 4.34TNALNHH408 pKa = 5.42LWEE411 pKa = 4.96RR412 pKa = 11.84LQSTHH417 pKa = 5.58SKK419 pKa = 10.04RR420 pKa = 11.84IKK422 pKa = 9.86RR423 pKa = 11.84SSNRR427 pKa = 11.84VKK429 pKa = 10.55RR430 pKa = 11.84APLGIIGRR438 pKa = 11.84LYY440 pKa = 10.13KK441 pKa = 10.06WLFGVATVDD450 pKa = 4.24DD451 pKa = 4.03VDD453 pKa = 3.56EE454 pKa = 4.14VRR456 pKa = 11.84RR457 pKa = 11.84NFVSYY462 pKa = 11.1LKK464 pKa = 10.62GQQNLSKK471 pKa = 10.81ANQEE475 pKa = 3.91FMTNQLSINQHH486 pKa = 5.13FDD488 pKa = 3.35EE489 pKa = 4.73NLKK492 pKa = 10.32ILNTKK497 pKa = 10.05ISDD500 pKa = 3.79TVKK503 pKa = 10.8GMNSTIDD510 pKa = 3.37MFMDD514 pKa = 5.39TIQQTNNVFQDD525 pKa = 3.97LYY527 pKa = 10.39LTTKK531 pKa = 10.61LSSLLIHH538 pKa = 6.4GNILITGAVIDD549 pKa = 4.21ALRR552 pKa = 11.84INQYY556 pKa = 9.37MDD558 pKa = 3.68SLSSRR563 pKa = 11.84YY564 pKa = 9.71LSLYY568 pKa = 9.86HH569 pKa = 7.18ALLSDD574 pKa = 3.71TLTAEE579 pKa = 4.14ILDD582 pKa = 4.14PASLKK587 pKa = 10.24TILLEE592 pKa = 4.52INNKK596 pKa = 9.62LPPNYY601 pKa = 9.77QVIDD605 pKa = 4.03LPDD608 pKa = 3.44FRR610 pKa = 11.84SWYY613 pKa = 8.42KK614 pKa = 10.35VSYY617 pKa = 9.07STVSLIDD624 pKa = 3.29NSIVYY629 pKa = 9.89AFKK632 pKa = 10.69FPIVRR637 pKa = 11.84KK638 pKa = 10.0SSEE641 pKa = 3.61PTLWHH646 pKa = 6.44FNSLPVFKK654 pKa = 10.86NGTAFKK660 pKa = 10.74LNYY663 pKa = 8.43HH664 pKa = 6.01DD665 pKa = 3.37HH666 pKa = 6.03WVYY669 pKa = 11.14LDD671 pKa = 3.38EE672 pKa = 6.27HH673 pKa = 5.37NQEE676 pKa = 4.33WVPMSQTDD684 pKa = 3.56FNARR688 pKa = 11.84CDD690 pKa = 3.59KK691 pKa = 10.14STSVCSGFASRR702 pKa = 11.84YY703 pKa = 7.2TYY705 pKa = 11.19SKK707 pKa = 11.31NNYY710 pKa = 6.96NHH712 pKa = 6.99CLLSLLYY719 pKa = 10.45DD720 pKa = 3.49SSKK723 pKa = 11.37VIDD726 pKa = 3.85QCEE729 pKa = 4.01MSPIDD734 pKa = 3.78IDD736 pKa = 3.93QVLDD740 pKa = 4.25PIIAISIPPNQWLVEE755 pKa = 3.84TLYY758 pKa = 10.28PVKK761 pKa = 10.36AVKK764 pKa = 10.16SCPAKK769 pKa = 10.3SGQFITDD776 pKa = 3.01SVMISGVMTIEE787 pKa = 4.58LAPLCKK793 pKa = 9.75ISIAEE798 pKa = 4.11LTLEE802 pKa = 4.61SSLAPPGLEE811 pKa = 4.0SQSYY815 pKa = 8.62APEE818 pKa = 3.94YY819 pKa = 9.39TLEE822 pKa = 4.14SVKK825 pKa = 10.52ISTSDD830 pKa = 2.85IDD832 pKa = 4.65IIALKK837 pKa = 9.56EE838 pKa = 4.11NKK840 pKa = 9.37VPQIDD845 pKa = 3.23IKK847 pKa = 10.38MDD849 pKa = 2.91KK850 pKa = 9.7STWNTKK856 pKa = 9.44AMQIKK861 pKa = 9.67IKK863 pKa = 10.72SVEE866 pKa = 4.27TNLNAQKK873 pKa = 10.54EE874 pKa = 4.41LVSNISEE881 pKa = 4.2INKK884 pKa = 9.59KK885 pKa = 9.89LQADD889 pKa = 4.76LIKK892 pKa = 10.34IEE894 pKa = 4.23SSSGKK899 pKa = 8.16TFSSQWISFYY909 pKa = 10.1WEE911 pKa = 3.91NIIFYY916 pKa = 9.15IWVIILTLMTLRR928 pKa = 11.84VYY930 pKa = 10.85VRR932 pKa = 11.84VFGIPLIGRR941 pKa = 11.84GNGIMAKK948 pKa = 8.96ATFPLVSSSGFGTANAFPPQGKK970 pKa = 9.22IYY972 pKa = 9.82MDD974 pKa = 4.74GSTIWASAPNATSNVMVNVTNKK996 pKa = 9.61FNEE999 pKa = 4.71FFSQPHH1005 pKa = 5.28NTTSVIHH1012 pKa = 5.97LCVMVALMLLMLILMYY1028 pKa = 11.19SLMCHH1033 pKa = 5.64HH1034 pKa = 6.91QSMLLRR1040 pKa = 11.84KK1041 pKa = 9.13LLSLGVLPINTRR1053 pKa = 11.84EE1054 pKa = 4.02SQHH1057 pKa = 7.4IGEE1060 pKa = 5.17IGVQLYY1066 pKa = 9.86VLVMMKK1072 pKa = 10.79SMFGSKK1078 pKa = 7.15THH1080 pKa = 5.45YY1081 pKa = 10.85AEE1083 pKa = 5.56VGIQMCTLPGNIRR1096 pKa = 11.84DD1097 pKa = 4.45WICTSNLSEE1106 pKa = 4.05NKK1108 pKa = 9.33TFIVPKK1114 pKa = 9.41WVCRR1118 pKa = 11.84WSSTLEE1124 pKa = 4.41LSCQWGPICLKK1135 pKa = 8.65STKK1138 pKa = 9.91YY1139 pKa = 10.71PNVDD1143 pKa = 3.08TCKK1146 pKa = 10.43DD1147 pKa = 3.72LPKK1150 pKa = 10.46HH1151 pKa = 5.97GSFNMRR1157 pKa = 11.84EE1158 pKa = 4.28VIEE1161 pKa = 4.33QINPKK1166 pKa = 10.34CPFIWKK1172 pKa = 9.44YY1173 pKa = 10.12ISAVEE1178 pKa = 4.05VNKK1181 pKa = 10.43VILRR1185 pKa = 11.84THH1187 pKa = 6.43NDD1189 pKa = 2.84YY1190 pKa = 11.47KK1191 pKa = 10.87EE1192 pKa = 4.1IYY1194 pKa = 9.53SYY1196 pKa = 11.79NSDD1199 pKa = 3.64TLSS1202 pKa = 3.13
Molecular weight: 136.4 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4334 |
673 |
2459 |
1444.7 |
164.86 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.73 ± 0.939 | 1.454 ± 0.205 |
5.653 ± 0.267 | 5.03 ± 0.207 |
4.199 ± 0.553 | 4.43 ± 0.33 |
2.653 ± 0.492 | 7.176 ± 0.155 |
5.699 ± 0.49 | 11.237 ± 0.828 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.276 ± 0.048 | 5.538 ± 0.597 |
4.522 ± 0.222 | 3.992 ± 0.678 |
4.569 ± 0.583 | 7.845 ± 0.633 |
6.714 ± 0.631 | 6.161 ± 0.212 |
1.777 ± 0.125 | 3.346 ± 0.405 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
