
Planctomicrobium piriforme
Taxonomy: cellular organisms; Bacteria; PVC group; Planctomycetes; Planctomycetia; Planctomycetales; Planctomycetaceae; Planctomicrobium
Average proteome isoelectric point is 6.34
Get precalculated fractions of proteins
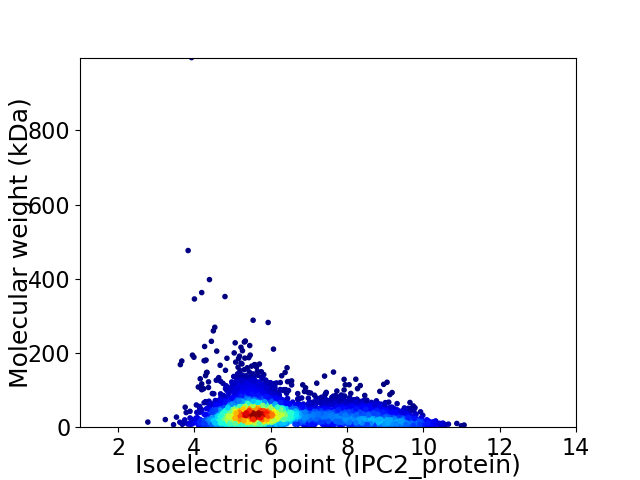
Virtual 2D-PAGE plot for 5057 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I3HHL5|A0A1I3HHL5_9PLAN Uncharacterized protein OS=Planctomicrobium piriforme OX=1576369 GN=SAMN05421753_10869 PE=4 SV=1
MM1 pKa = 7.34LRR3 pKa = 11.84KK4 pKa = 9.68SLSSLLWSAWGRR16 pKa = 11.84LQAGSRR22 pKa = 11.84NSGRR26 pKa = 11.84GRR28 pKa = 11.84RR29 pKa = 11.84PLQRR33 pKa = 11.84HH34 pKa = 4.06SWNRR38 pKa = 11.84SLSRR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84QSGVPALEE53 pKa = 3.89RR54 pKa = 11.84LEE56 pKa = 4.59ARR58 pKa = 11.84QLLSATASGAEE69 pKa = 4.02YY70 pKa = 10.25RR71 pKa = 11.84VNAYY75 pKa = 6.29TTSIQSQPSMAMDD88 pKa = 3.45SAGNYY93 pKa = 7.71VVTWQSYY100 pKa = 8.16GQDD103 pKa = 3.12GDD105 pKa = 4.03GYY107 pKa = 10.94GIYY110 pKa = 10.35AQRR113 pKa = 11.84FNAAGVAQGSEE124 pKa = 4.26FRR126 pKa = 11.84VNTYY130 pKa = 5.47TTYY133 pKa = 10.88YY134 pKa = 10.01QFNPSAAMDD143 pKa = 3.75SDD145 pKa = 3.88GDD147 pKa = 4.54FVITWVTDD155 pKa = 3.47QEE157 pKa = 4.69GSSNDD162 pKa = 3.04IYY164 pKa = 10.71AQRR167 pKa = 11.84YY168 pKa = 5.85NALGEE173 pKa = 4.1AQGTEE178 pKa = 4.16FRR180 pKa = 11.84VNTYY184 pKa = 7.54TTDD187 pKa = 3.45PQYY190 pKa = 11.45SPSAAMDD197 pKa = 3.51SDD199 pKa = 3.76GDD201 pKa = 4.95FVVTWQSYY209 pKa = 8.32GQDD212 pKa = 3.12GDD214 pKa = 4.03GYY216 pKa = 10.94GIYY219 pKa = 10.35AQRR222 pKa = 11.84FNAAGVAQGSEE233 pKa = 4.37FQVNTYY239 pKa = 6.31TTSYY243 pKa = 10.78QYY245 pKa = 9.89TASAAMDD252 pKa = 3.68SDD254 pKa = 3.92GDD256 pKa = 5.21FVITWQSYY264 pKa = 7.9GQDD267 pKa = 2.97GSGYY271 pKa = 9.91GIYY274 pKa = 9.89AQRR277 pKa = 11.84YY278 pKa = 5.86NAAGVAQGSEE288 pKa = 4.44FQVSSYY294 pKa = 7.25TTNNQNSSSAAMDD307 pKa = 3.43SDD309 pKa = 3.84GDD311 pKa = 4.86FVISWQSSGQDD322 pKa = 3.16GSGYY326 pKa = 10.49GVYY329 pKa = 10.57AKK331 pKa = 10.21RR332 pKa = 11.84FNASGVAQGSEE343 pKa = 4.18FRR345 pKa = 11.84VNTYY349 pKa = 7.27TTSSQYY355 pKa = 10.51NASPAMNAAGNFVISWQSSGQDD377 pKa = 3.49GNGLGIYY384 pKa = 8.55AQYY387 pKa = 11.04YY388 pKa = 7.9NAAGVAQGSEE398 pKa = 4.35FRR400 pKa = 11.84ANSYY404 pKa = 6.77TTNSQANPTAAMNSTGNFVIAWQSSGQDD432 pKa = 3.27GSSNGIYY439 pKa = 8.55SQRR442 pKa = 11.84YY443 pKa = 8.22SVGAPPTDD451 pKa = 3.02ISLSANTVSEE461 pKa = 4.11IASIGTTIGTLTATDD476 pKa = 3.79ADD478 pKa = 4.05SPEE481 pKa = 4.16SFTYY485 pKa = 10.5RR486 pKa = 11.84LLTNVEE492 pKa = 4.42SPANGLFLIDD502 pKa = 4.38GDD504 pKa = 4.28TLKK507 pKa = 10.07TAGALNYY514 pKa = 9.24EE515 pKa = 4.37AVHH518 pKa = 5.97QYY520 pKa = 10.99SVDD523 pKa = 3.42IEE525 pKa = 4.64VTDD528 pKa = 3.67STAQTFVKK536 pKa = 9.8TFTITVTDD544 pKa = 3.79SNEE547 pKa = 3.92FAVSAVSDD555 pKa = 3.96TNGDD559 pKa = 3.61TNTIAEE565 pKa = 4.16NAAAGTVGITASASDD580 pKa = 4.28ADD582 pKa = 3.97GTTNAVSYY590 pKa = 11.09SLTDD594 pKa = 3.29NAGGLFTIDD603 pKa = 2.99STTGVVSTTGPLNYY617 pKa = 7.74EE618 pKa = 4.02TATSYY623 pKa = 10.77TITVRR628 pKa = 11.84AEE630 pKa = 3.89SADD633 pKa = 3.62GSFSAADD640 pKa = 3.41FTITVTDD647 pKa = 3.35IDD649 pKa = 4.32EE650 pKa = 4.92SDD652 pKa = 3.53VSPITDD658 pKa = 3.44SDD660 pKa = 4.07SASNTIAEE668 pKa = 4.18NAAAGTVGITASASDD683 pKa = 4.18ADD685 pKa = 4.02GTTNTVTYY693 pKa = 10.72SLTDD697 pKa = 3.28NAGGLFTIDD706 pKa = 2.98SATGVVSTTGPLDD719 pKa = 3.46YY720 pKa = 9.48EE721 pKa = 4.51TATSYY726 pKa = 11.12SITVRR731 pKa = 11.84AEE733 pKa = 3.83SADD736 pKa = 3.6GSFSTADD743 pKa = 3.17FTIAVTDD750 pKa = 3.48VDD752 pKa = 4.0EE753 pKa = 5.68SDD755 pKa = 3.5VSSITDD761 pKa = 3.19TDD763 pKa = 3.81TADD766 pKa = 3.39NTITEE771 pKa = 4.34SADD774 pKa = 3.05AGTVGITASASDD786 pKa = 4.18ADD788 pKa = 4.02GTTNTVTYY796 pKa = 10.72SLTDD800 pKa = 3.28NAGGLFTIDD809 pKa = 2.99STTGVVSTTGPLNYY823 pKa = 7.74EE824 pKa = 4.02TATSYY829 pKa = 10.77TITVRR834 pKa = 11.84AEE836 pKa = 3.99SEE838 pKa = 4.17DD839 pKa = 3.84GSFSAADD846 pKa = 3.51FTVTVTDD853 pKa = 3.22VDD855 pKa = 3.87EE856 pKa = 5.7SDD858 pKa = 3.59VSPITDD864 pKa = 3.44SDD866 pKa = 4.07SASNTIAEE874 pKa = 4.32NADD877 pKa = 3.32AGTVGITASASDD889 pKa = 4.18ADD891 pKa = 4.02GTTNTVTYY899 pKa = 10.72SLTDD903 pKa = 3.28NAGGLFTIDD912 pKa = 2.99STTGVVSTTGPLNYY926 pKa = 7.74EE927 pKa = 4.02TATSYY932 pKa = 10.77TITVRR937 pKa = 11.84AEE939 pKa = 3.99SEE941 pKa = 4.15DD942 pKa = 3.8GSFSTTDD949 pKa = 3.28FTIAVTDD956 pKa = 3.48VDD958 pKa = 3.91EE959 pKa = 5.53SDD961 pKa = 3.43VSGVTDD967 pKa = 3.75TDD969 pKa = 3.7STDD972 pKa = 3.12NTISEE977 pKa = 4.15NAAAGPVGITAFASDD992 pKa = 4.78ADD994 pKa = 3.96GTTNAVSYY1002 pKa = 11.09SLTDD1006 pKa = 3.29NAGGLFTIDD1015 pKa = 3.95SISGVVSTTGPLDD1028 pKa = 3.46YY1029 pKa = 9.48EE1030 pKa = 4.51TATSYY1035 pKa = 11.12SITVRR1040 pKa = 11.84AEE1042 pKa = 3.83SADD1045 pKa = 3.48GSFSIADD1052 pKa = 3.26FTIAVTDD1059 pKa = 3.31VDD1061 pKa = 4.33EE1062 pKa = 5.36FDD1064 pKa = 3.68VSAIADD1070 pKa = 3.52ADD1072 pKa = 3.92AADD1075 pKa = 3.62NTIAEE1080 pKa = 4.45NADD1083 pKa = 3.59AGPVGITASASDD1095 pKa = 3.76SDD1097 pKa = 4.17GATNTVTYY1105 pKa = 10.86SLTDD1109 pKa = 3.48DD1110 pKa = 3.84AGGLFTIDD1118 pKa = 3.95SISGVVSTTGPLNYY1132 pKa = 7.7EE1133 pKa = 4.02TATSYY1138 pKa = 11.1SITVRR1143 pKa = 11.84AEE1145 pKa = 3.83SADD1148 pKa = 3.31GSFSTEE1154 pKa = 3.47NFTIAVTDD1162 pKa = 3.55VNEE1165 pKa = 4.09FDD1167 pKa = 3.63VSDD1170 pKa = 4.93VIDD1173 pKa = 3.9TNCASNVIAEE1183 pKa = 4.22NAAAGPIGITAFADD1197 pKa = 4.13DD1198 pKa = 4.94EE1199 pKa = 5.2DD1200 pKa = 4.49GTTNGVTYY1208 pKa = 10.88SLTDD1212 pKa = 3.47DD1213 pKa = 3.84AGGLFTINSCTGVVSTTGPLDD1234 pKa = 3.4YY1235 pKa = 9.46EE1236 pKa = 4.7TACSYY1241 pKa = 11.03SITVRR1246 pKa = 11.84AEE1248 pKa = 3.83SADD1251 pKa = 3.5GSFSMADD1258 pKa = 3.19FTVNVLNVNEE1268 pKa = 4.45SPVISGVTGSTSYY1281 pKa = 10.86IEE1283 pKa = 4.03NAAPRR1288 pKa = 11.84ILTTDD1293 pKa = 3.51GVINDD1298 pKa = 4.51PDD1300 pKa = 4.09GNFCGGSLTVRR1311 pKa = 11.84FRR1313 pKa = 11.84GFSEE1317 pKa = 3.85TGDD1320 pKa = 3.49RR1321 pKa = 11.84LSILSVGTSTGQISVTGNDD1340 pKa = 2.98VFYY1343 pKa = 11.1GCTLLGTVTSDD1354 pKa = 3.06GTGGQNLVIQLAAGADD1370 pKa = 3.61NAGVQQLLRR1379 pKa = 11.84SITYY1383 pKa = 10.31ASVSEE1388 pKa = 4.4NPSTTQRR1395 pKa = 11.84RR1396 pKa = 11.84VEE1398 pKa = 4.1FVLKK1402 pKa = 10.74DD1403 pKa = 3.78GVGTASNVGAKK1414 pKa = 8.29VVRR1417 pKa = 11.84VTAVNDD1423 pKa = 3.52KK1424 pKa = 10.64PVITGVTGSTTYY1436 pKa = 10.44TEE1438 pKa = 4.01NAAPLVLATDD1448 pKa = 4.72GVISDD1453 pKa = 4.81LDD1455 pKa = 4.12DD1456 pKa = 3.95NLGEE1460 pKa = 4.28GSLTVRR1466 pKa = 11.84FNGFSEE1472 pKa = 4.47ADD1474 pKa = 3.43DD1475 pKa = 3.88RR1476 pKa = 11.84LSILSVGTGDD1486 pKa = 3.68GQISVTGNDD1495 pKa = 3.0VFYY1498 pKa = 10.94GGSLLGTISSDD1509 pKa = 3.04GTGGQNLVIQLASGASSEE1527 pKa = 5.05AVQQLLRR1534 pKa = 11.84SITYY1538 pKa = 10.31ASVSEE1543 pKa = 4.38NPNTTQRR1550 pKa = 11.84RR1551 pKa = 11.84VEE1553 pKa = 3.82FALRR1557 pKa = 11.84DD1558 pKa = 3.48GDD1560 pKa = 4.03GANSNVGARR1569 pKa = 11.84IVKK1572 pKa = 9.54VKK1574 pKa = 10.51SVNDD1578 pKa = 3.55KK1579 pKa = 10.76PVITGVTGTATYY1591 pKa = 10.26SKK1593 pKa = 10.54SGLPIALATGGDD1605 pKa = 3.84VSDD1608 pKa = 5.77LDD1610 pKa = 3.66WNMAGGSLTARR1621 pKa = 11.84FNGFSEE1627 pKa = 4.58ANDD1630 pKa = 3.63RR1631 pKa = 11.84LSIISGGTGTGEE1643 pKa = 3.97INVSGNDD1650 pKa = 3.2VYY1652 pKa = 11.82YY1653 pKa = 10.5EE1654 pKa = 4.23GDD1656 pKa = 4.05LIGSITSNGIGGQALTITLTAAATPDD1682 pKa = 3.51AVRR1685 pKa = 11.84HH1686 pKa = 5.6LLHH1689 pKa = 6.52SLTYY1693 pKa = 10.57ASDD1696 pKa = 3.44STTPTTTNRR1705 pKa = 11.84RR1706 pKa = 11.84VEE1708 pKa = 4.13FLLTDD1713 pKa = 3.61SSGAKK1718 pKa = 10.05SNTAARR1724 pKa = 11.84VVKK1727 pKa = 10.15FVSS1730 pKa = 3.15
MM1 pKa = 7.34LRR3 pKa = 11.84KK4 pKa = 9.68SLSSLLWSAWGRR16 pKa = 11.84LQAGSRR22 pKa = 11.84NSGRR26 pKa = 11.84GRR28 pKa = 11.84RR29 pKa = 11.84PLQRR33 pKa = 11.84HH34 pKa = 4.06SWNRR38 pKa = 11.84SLSRR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84QSGVPALEE53 pKa = 3.89RR54 pKa = 11.84LEE56 pKa = 4.59ARR58 pKa = 11.84QLLSATASGAEE69 pKa = 4.02YY70 pKa = 10.25RR71 pKa = 11.84VNAYY75 pKa = 6.29TTSIQSQPSMAMDD88 pKa = 3.45SAGNYY93 pKa = 7.71VVTWQSYY100 pKa = 8.16GQDD103 pKa = 3.12GDD105 pKa = 4.03GYY107 pKa = 10.94GIYY110 pKa = 10.35AQRR113 pKa = 11.84FNAAGVAQGSEE124 pKa = 4.26FRR126 pKa = 11.84VNTYY130 pKa = 5.47TTYY133 pKa = 10.88YY134 pKa = 10.01QFNPSAAMDD143 pKa = 3.75SDD145 pKa = 3.88GDD147 pKa = 4.54FVITWVTDD155 pKa = 3.47QEE157 pKa = 4.69GSSNDD162 pKa = 3.04IYY164 pKa = 10.71AQRR167 pKa = 11.84YY168 pKa = 5.85NALGEE173 pKa = 4.1AQGTEE178 pKa = 4.16FRR180 pKa = 11.84VNTYY184 pKa = 7.54TTDD187 pKa = 3.45PQYY190 pKa = 11.45SPSAAMDD197 pKa = 3.51SDD199 pKa = 3.76GDD201 pKa = 4.95FVVTWQSYY209 pKa = 8.32GQDD212 pKa = 3.12GDD214 pKa = 4.03GYY216 pKa = 10.94GIYY219 pKa = 10.35AQRR222 pKa = 11.84FNAAGVAQGSEE233 pKa = 4.37FQVNTYY239 pKa = 6.31TTSYY243 pKa = 10.78QYY245 pKa = 9.89TASAAMDD252 pKa = 3.68SDD254 pKa = 3.92GDD256 pKa = 5.21FVITWQSYY264 pKa = 7.9GQDD267 pKa = 2.97GSGYY271 pKa = 9.91GIYY274 pKa = 9.89AQRR277 pKa = 11.84YY278 pKa = 5.86NAAGVAQGSEE288 pKa = 4.44FQVSSYY294 pKa = 7.25TTNNQNSSSAAMDD307 pKa = 3.43SDD309 pKa = 3.84GDD311 pKa = 4.86FVISWQSSGQDD322 pKa = 3.16GSGYY326 pKa = 10.49GVYY329 pKa = 10.57AKK331 pKa = 10.21RR332 pKa = 11.84FNASGVAQGSEE343 pKa = 4.18FRR345 pKa = 11.84VNTYY349 pKa = 7.27TTSSQYY355 pKa = 10.51NASPAMNAAGNFVISWQSSGQDD377 pKa = 3.49GNGLGIYY384 pKa = 8.55AQYY387 pKa = 11.04YY388 pKa = 7.9NAAGVAQGSEE398 pKa = 4.35FRR400 pKa = 11.84ANSYY404 pKa = 6.77TTNSQANPTAAMNSTGNFVIAWQSSGQDD432 pKa = 3.27GSSNGIYY439 pKa = 8.55SQRR442 pKa = 11.84YY443 pKa = 8.22SVGAPPTDD451 pKa = 3.02ISLSANTVSEE461 pKa = 4.11IASIGTTIGTLTATDD476 pKa = 3.79ADD478 pKa = 4.05SPEE481 pKa = 4.16SFTYY485 pKa = 10.5RR486 pKa = 11.84LLTNVEE492 pKa = 4.42SPANGLFLIDD502 pKa = 4.38GDD504 pKa = 4.28TLKK507 pKa = 10.07TAGALNYY514 pKa = 9.24EE515 pKa = 4.37AVHH518 pKa = 5.97QYY520 pKa = 10.99SVDD523 pKa = 3.42IEE525 pKa = 4.64VTDD528 pKa = 3.67STAQTFVKK536 pKa = 9.8TFTITVTDD544 pKa = 3.79SNEE547 pKa = 3.92FAVSAVSDD555 pKa = 3.96TNGDD559 pKa = 3.61TNTIAEE565 pKa = 4.16NAAAGTVGITASASDD580 pKa = 4.28ADD582 pKa = 3.97GTTNAVSYY590 pKa = 11.09SLTDD594 pKa = 3.29NAGGLFTIDD603 pKa = 2.99STTGVVSTTGPLNYY617 pKa = 7.74EE618 pKa = 4.02TATSYY623 pKa = 10.77TITVRR628 pKa = 11.84AEE630 pKa = 3.89SADD633 pKa = 3.62GSFSAADD640 pKa = 3.41FTITVTDD647 pKa = 3.35IDD649 pKa = 4.32EE650 pKa = 4.92SDD652 pKa = 3.53VSPITDD658 pKa = 3.44SDD660 pKa = 4.07SASNTIAEE668 pKa = 4.18NAAAGTVGITASASDD683 pKa = 4.18ADD685 pKa = 4.02GTTNTVTYY693 pKa = 10.72SLTDD697 pKa = 3.28NAGGLFTIDD706 pKa = 2.98SATGVVSTTGPLDD719 pKa = 3.46YY720 pKa = 9.48EE721 pKa = 4.51TATSYY726 pKa = 11.12SITVRR731 pKa = 11.84AEE733 pKa = 3.83SADD736 pKa = 3.6GSFSTADD743 pKa = 3.17FTIAVTDD750 pKa = 3.48VDD752 pKa = 4.0EE753 pKa = 5.68SDD755 pKa = 3.5VSSITDD761 pKa = 3.19TDD763 pKa = 3.81TADD766 pKa = 3.39NTITEE771 pKa = 4.34SADD774 pKa = 3.05AGTVGITASASDD786 pKa = 4.18ADD788 pKa = 4.02GTTNTVTYY796 pKa = 10.72SLTDD800 pKa = 3.28NAGGLFTIDD809 pKa = 2.99STTGVVSTTGPLNYY823 pKa = 7.74EE824 pKa = 4.02TATSYY829 pKa = 10.77TITVRR834 pKa = 11.84AEE836 pKa = 3.99SEE838 pKa = 4.17DD839 pKa = 3.84GSFSAADD846 pKa = 3.51FTVTVTDD853 pKa = 3.22VDD855 pKa = 3.87EE856 pKa = 5.7SDD858 pKa = 3.59VSPITDD864 pKa = 3.44SDD866 pKa = 4.07SASNTIAEE874 pKa = 4.32NADD877 pKa = 3.32AGTVGITASASDD889 pKa = 4.18ADD891 pKa = 4.02GTTNTVTYY899 pKa = 10.72SLTDD903 pKa = 3.28NAGGLFTIDD912 pKa = 2.99STTGVVSTTGPLNYY926 pKa = 7.74EE927 pKa = 4.02TATSYY932 pKa = 10.77TITVRR937 pKa = 11.84AEE939 pKa = 3.99SEE941 pKa = 4.15DD942 pKa = 3.8GSFSTTDD949 pKa = 3.28FTIAVTDD956 pKa = 3.48VDD958 pKa = 3.91EE959 pKa = 5.53SDD961 pKa = 3.43VSGVTDD967 pKa = 3.75TDD969 pKa = 3.7STDD972 pKa = 3.12NTISEE977 pKa = 4.15NAAAGPVGITAFASDD992 pKa = 4.78ADD994 pKa = 3.96GTTNAVSYY1002 pKa = 11.09SLTDD1006 pKa = 3.29NAGGLFTIDD1015 pKa = 3.95SISGVVSTTGPLDD1028 pKa = 3.46YY1029 pKa = 9.48EE1030 pKa = 4.51TATSYY1035 pKa = 11.12SITVRR1040 pKa = 11.84AEE1042 pKa = 3.83SADD1045 pKa = 3.48GSFSIADD1052 pKa = 3.26FTIAVTDD1059 pKa = 3.31VDD1061 pKa = 4.33EE1062 pKa = 5.36FDD1064 pKa = 3.68VSAIADD1070 pKa = 3.52ADD1072 pKa = 3.92AADD1075 pKa = 3.62NTIAEE1080 pKa = 4.45NADD1083 pKa = 3.59AGPVGITASASDD1095 pKa = 3.76SDD1097 pKa = 4.17GATNTVTYY1105 pKa = 10.86SLTDD1109 pKa = 3.48DD1110 pKa = 3.84AGGLFTIDD1118 pKa = 3.95SISGVVSTTGPLNYY1132 pKa = 7.7EE1133 pKa = 4.02TATSYY1138 pKa = 11.1SITVRR1143 pKa = 11.84AEE1145 pKa = 3.83SADD1148 pKa = 3.31GSFSTEE1154 pKa = 3.47NFTIAVTDD1162 pKa = 3.55VNEE1165 pKa = 4.09FDD1167 pKa = 3.63VSDD1170 pKa = 4.93VIDD1173 pKa = 3.9TNCASNVIAEE1183 pKa = 4.22NAAAGPIGITAFADD1197 pKa = 4.13DD1198 pKa = 4.94EE1199 pKa = 5.2DD1200 pKa = 4.49GTTNGVTYY1208 pKa = 10.88SLTDD1212 pKa = 3.47DD1213 pKa = 3.84AGGLFTINSCTGVVSTTGPLDD1234 pKa = 3.4YY1235 pKa = 9.46EE1236 pKa = 4.7TACSYY1241 pKa = 11.03SITVRR1246 pKa = 11.84AEE1248 pKa = 3.83SADD1251 pKa = 3.5GSFSMADD1258 pKa = 3.19FTVNVLNVNEE1268 pKa = 4.45SPVISGVTGSTSYY1281 pKa = 10.86IEE1283 pKa = 4.03NAAPRR1288 pKa = 11.84ILTTDD1293 pKa = 3.51GVINDD1298 pKa = 4.51PDD1300 pKa = 4.09GNFCGGSLTVRR1311 pKa = 11.84FRR1313 pKa = 11.84GFSEE1317 pKa = 3.85TGDD1320 pKa = 3.49RR1321 pKa = 11.84LSILSVGTSTGQISVTGNDD1340 pKa = 2.98VFYY1343 pKa = 11.1GCTLLGTVTSDD1354 pKa = 3.06GTGGQNLVIQLAAGADD1370 pKa = 3.61NAGVQQLLRR1379 pKa = 11.84SITYY1383 pKa = 10.31ASVSEE1388 pKa = 4.4NPSTTQRR1395 pKa = 11.84RR1396 pKa = 11.84VEE1398 pKa = 4.1FVLKK1402 pKa = 10.74DD1403 pKa = 3.78GVGTASNVGAKK1414 pKa = 8.29VVRR1417 pKa = 11.84VTAVNDD1423 pKa = 3.52KK1424 pKa = 10.64PVITGVTGSTTYY1436 pKa = 10.44TEE1438 pKa = 4.01NAAPLVLATDD1448 pKa = 4.72GVISDD1453 pKa = 4.81LDD1455 pKa = 4.12DD1456 pKa = 3.95NLGEE1460 pKa = 4.28GSLTVRR1466 pKa = 11.84FNGFSEE1472 pKa = 4.47ADD1474 pKa = 3.43DD1475 pKa = 3.88RR1476 pKa = 11.84LSILSVGTGDD1486 pKa = 3.68GQISVTGNDD1495 pKa = 3.0VFYY1498 pKa = 10.94GGSLLGTISSDD1509 pKa = 3.04GTGGQNLVIQLASGASSEE1527 pKa = 5.05AVQQLLRR1534 pKa = 11.84SITYY1538 pKa = 10.31ASVSEE1543 pKa = 4.38NPNTTQRR1550 pKa = 11.84RR1551 pKa = 11.84VEE1553 pKa = 3.82FALRR1557 pKa = 11.84DD1558 pKa = 3.48GDD1560 pKa = 4.03GANSNVGARR1569 pKa = 11.84IVKK1572 pKa = 9.54VKK1574 pKa = 10.51SVNDD1578 pKa = 3.55KK1579 pKa = 10.76PVITGVTGTATYY1591 pKa = 10.26SKK1593 pKa = 10.54SGLPIALATGGDD1605 pKa = 3.84VSDD1608 pKa = 5.77LDD1610 pKa = 3.66WNMAGGSLTARR1621 pKa = 11.84FNGFSEE1627 pKa = 4.58ANDD1630 pKa = 3.63RR1631 pKa = 11.84LSIISGGTGTGEE1643 pKa = 3.97INVSGNDD1650 pKa = 3.2VYY1652 pKa = 11.82YY1653 pKa = 10.5EE1654 pKa = 4.23GDD1656 pKa = 4.05LIGSITSNGIGGQALTITLTAAATPDD1682 pKa = 3.51AVRR1685 pKa = 11.84HH1686 pKa = 5.6LLHH1689 pKa = 6.52SLTYY1693 pKa = 10.57ASDD1696 pKa = 3.44STTPTTTNRR1705 pKa = 11.84RR1706 pKa = 11.84VEE1708 pKa = 4.13FLLTDD1713 pKa = 3.61SSGAKK1718 pKa = 10.05SNTAARR1724 pKa = 11.84VVKK1727 pKa = 10.15FVSS1730 pKa = 3.15
Molecular weight: 178.39 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I3LYJ1|A0A1I3LYJ1_9PLAN Putative signal transducing protein OS=Planctomicrobium piriforme OX=1576369 GN=SAMN05421753_113106 PE=4 SV=1
MM1 pKa = 7.66AVILLIALMSNRR13 pKa = 11.84QPSMQQSSSVRR24 pKa = 11.84STSAQTVSNRR34 pKa = 11.84NAQTQRR40 pKa = 11.84GPLRR44 pKa = 11.84RR45 pKa = 11.84LVGRR49 pKa = 11.84MRR51 pKa = 11.84NN52 pKa = 3.38
MM1 pKa = 7.66AVILLIALMSNRR13 pKa = 11.84QPSMQQSSSVRR24 pKa = 11.84STSAQTVSNRR34 pKa = 11.84NAQTQRR40 pKa = 11.84GPLRR44 pKa = 11.84RR45 pKa = 11.84LVGRR49 pKa = 11.84MRR51 pKa = 11.84NN52 pKa = 3.38
Molecular weight: 5.8 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1801768 |
27 |
9591 |
356.3 |
39.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.904 ± 0.037 | 1.18 ± 0.015 |
5.482 ± 0.029 | 5.907 ± 0.038 |
3.803 ± 0.023 | 7.752 ± 0.044 |
2.136 ± 0.016 | 4.895 ± 0.023 |
3.673 ± 0.033 | 10.345 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.135 ± 0.017 | 3.209 ± 0.031 |
5.522 ± 0.031 | 4.446 ± 0.026 |
6.463 ± 0.043 | 6.282 ± 0.036 |
5.694 ± 0.054 | 7.24 ± 0.029 |
1.543 ± 0.017 | 2.387 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
