
Saprospira grandis (strain Lewin)
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Saprospiria; Saprospirales; Saprospiraceae; Saprospira; Saprospira grandis
Average proteome isoelectric point is 6.91
Get precalculated fractions of proteins
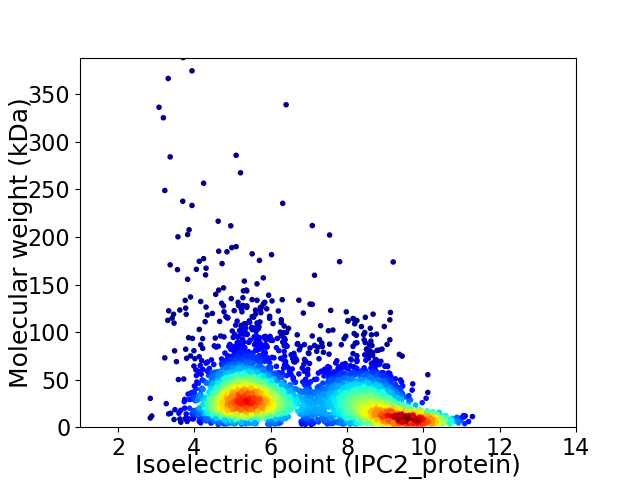
Virtual 2D-PAGE plot for 4218 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|H6KZG7|H6KZG7_SAPGL Membrane protein OS=Saprospira grandis (strain Lewin) OX=984262 GN=SGRA_3015 PE=4 SV=1
MM1 pKa = 7.54FNQKK5 pKa = 8.05LTQVLVALAFLLSAGSLSAQTVLWGAGSSDD35 pKa = 3.58TQIDD39 pKa = 4.04SIGRR43 pKa = 11.84FATDD47 pKa = 3.78TIIGLDD53 pKa = 3.63GLGWTVDD60 pKa = 3.49SVAGKK65 pKa = 9.31WEE67 pKa = 3.87YY68 pKa = 11.25SVDD71 pKa = 4.32GISQGTFSSGTPGLTSPSVGDD92 pKa = 3.52GVAFFDD98 pKa = 3.95ADD100 pKa = 3.31FWFAQGANPQIGTITSPAFDD120 pKa = 3.39LTGYY124 pKa = 9.73EE125 pKa = 4.01DD126 pKa = 4.18SIVVLEE132 pKa = 5.34FYY134 pKa = 11.0AGLRR138 pKa = 11.84DD139 pKa = 4.78FRR141 pKa = 11.84STDD144 pKa = 2.9NSAGFSTDD152 pKa = 5.0GGATWTDD159 pKa = 3.68FDD161 pKa = 4.63VISAIGAPSGSAGEE175 pKa = 4.2GMVLFDD181 pKa = 5.29LSNIVNSATSLTNCHH196 pKa = 5.2VRR198 pKa = 11.84FSFEE202 pKa = 3.61GDD204 pKa = 2.95SYY206 pKa = 11.61YY207 pKa = 11.25YY208 pKa = 10.92GVDD211 pKa = 3.29DD212 pKa = 3.75VTVRR216 pKa = 11.84TSEE219 pKa = 4.46AIYY222 pKa = 10.48DD223 pKa = 3.53IAIDD227 pKa = 3.85EE228 pKa = 4.76NYY230 pKa = 10.55NPPKK234 pKa = 10.71VVGSKK239 pKa = 9.49QLPLRR244 pKa = 11.84MVNADD249 pKa = 3.36EE250 pKa = 4.72MGWGANIEE258 pKa = 4.21NNGIGGNINAGEE270 pKa = 4.03AMLYY274 pKa = 8.33VTVSEE279 pKa = 4.51VSSGTVAVMDD289 pKa = 4.24SMAVPAIAPGTDD301 pKa = 2.77TALYY305 pKa = 9.47MPGFSNGWMPTAAGDD320 pKa = 3.67YY321 pKa = 9.99RR322 pKa = 11.84VDD324 pKa = 3.54YY325 pKa = 10.65SVSYY329 pKa = 9.28SANMQIDD336 pKa = 3.9TTNDD340 pKa = 2.68VYY342 pKa = 11.55TDD344 pKa = 4.23FFTITADD351 pKa = 3.72DD352 pKa = 4.03YY353 pKa = 11.09LSKK356 pKa = 10.76VGRR359 pKa = 11.84DD360 pKa = 3.25LTGFPSADD368 pKa = 3.25AATLPGVRR376 pKa = 11.84AADD379 pKa = 3.41GFLPAEE385 pKa = 4.47HH386 pKa = 7.0EE387 pKa = 4.37WGSMYY392 pKa = 10.31YY393 pKa = 9.68IPNYY397 pKa = 9.42TVTNGDD403 pKa = 4.11SIVADD408 pKa = 3.35SFLYY412 pKa = 10.25TGATVNADD420 pKa = 3.5TSVTEE425 pKa = 3.9AVVQVRR431 pKa = 11.84LYY433 pKa = 10.97KK434 pKa = 10.95FNDD437 pKa = 3.51DD438 pKa = 4.51DD439 pKa = 6.38GSGFWTAQPTGPSDD453 pKa = 3.73PEE455 pKa = 4.15LPIQALGVDD464 pKa = 4.04TFEE467 pKa = 6.34LDD469 pKa = 2.85LSLAFRR475 pKa = 11.84QRR477 pKa = 11.84AVALSNLSGGTGAVLQPNSFYY498 pKa = 11.19VATVLCSDD506 pKa = 4.58QINGLRR512 pKa = 11.84NADD515 pKa = 3.15NTTRR519 pKa = 11.84HH520 pKa = 5.39IFIGNSEE527 pKa = 4.45DD528 pKa = 4.74KK529 pKa = 11.19YY530 pKa = 11.58DD531 pKa = 3.73FTIDD535 pKa = 3.38SLPNYY540 pKa = 7.64FIPASIVRR548 pKa = 11.84SAYY551 pKa = 9.3IDD553 pKa = 3.33PTGTVTGLQDD563 pKa = 3.73NQWYY567 pKa = 8.39GAGYY571 pKa = 9.98EE572 pKa = 4.15SGIVPSIGLTLTDD585 pKa = 5.55AITINVTANEE595 pKa = 3.99AVTTEE600 pKa = 4.41FNIFPNPATEE610 pKa = 4.05QISAKK615 pKa = 10.52VEE617 pKa = 3.95LTEE620 pKa = 4.02MNSNVEE626 pKa = 4.13YY627 pKa = 10.33IITDD631 pKa = 3.08ATGRR635 pKa = 11.84VVIKK639 pKa = 8.8EE640 pKa = 4.01QKK642 pKa = 10.67ADD644 pKa = 3.67LQSDD648 pKa = 4.2VVTYY652 pKa = 10.52DD653 pKa = 3.19VSQLPAGVYY662 pKa = 9.43FFTVRR667 pKa = 11.84TAQGASSQKK676 pKa = 9.9FVKK679 pKa = 10.06QQ680 pKa = 3.29
MM1 pKa = 7.54FNQKK5 pKa = 8.05LTQVLVALAFLLSAGSLSAQTVLWGAGSSDD35 pKa = 3.58TQIDD39 pKa = 4.04SIGRR43 pKa = 11.84FATDD47 pKa = 3.78TIIGLDD53 pKa = 3.63GLGWTVDD60 pKa = 3.49SVAGKK65 pKa = 9.31WEE67 pKa = 3.87YY68 pKa = 11.25SVDD71 pKa = 4.32GISQGTFSSGTPGLTSPSVGDD92 pKa = 3.52GVAFFDD98 pKa = 3.95ADD100 pKa = 3.31FWFAQGANPQIGTITSPAFDD120 pKa = 3.39LTGYY124 pKa = 9.73EE125 pKa = 4.01DD126 pKa = 4.18SIVVLEE132 pKa = 5.34FYY134 pKa = 11.0AGLRR138 pKa = 11.84DD139 pKa = 4.78FRR141 pKa = 11.84STDD144 pKa = 2.9NSAGFSTDD152 pKa = 5.0GGATWTDD159 pKa = 3.68FDD161 pKa = 4.63VISAIGAPSGSAGEE175 pKa = 4.2GMVLFDD181 pKa = 5.29LSNIVNSATSLTNCHH196 pKa = 5.2VRR198 pKa = 11.84FSFEE202 pKa = 3.61GDD204 pKa = 2.95SYY206 pKa = 11.61YY207 pKa = 11.25YY208 pKa = 10.92GVDD211 pKa = 3.29DD212 pKa = 3.75VTVRR216 pKa = 11.84TSEE219 pKa = 4.46AIYY222 pKa = 10.48DD223 pKa = 3.53IAIDD227 pKa = 3.85EE228 pKa = 4.76NYY230 pKa = 10.55NPPKK234 pKa = 10.71VVGSKK239 pKa = 9.49QLPLRR244 pKa = 11.84MVNADD249 pKa = 3.36EE250 pKa = 4.72MGWGANIEE258 pKa = 4.21NNGIGGNINAGEE270 pKa = 4.03AMLYY274 pKa = 8.33VTVSEE279 pKa = 4.51VSSGTVAVMDD289 pKa = 4.24SMAVPAIAPGTDD301 pKa = 2.77TALYY305 pKa = 9.47MPGFSNGWMPTAAGDD320 pKa = 3.67YY321 pKa = 9.99RR322 pKa = 11.84VDD324 pKa = 3.54YY325 pKa = 10.65SVSYY329 pKa = 9.28SANMQIDD336 pKa = 3.9TTNDD340 pKa = 2.68VYY342 pKa = 11.55TDD344 pKa = 4.23FFTITADD351 pKa = 3.72DD352 pKa = 4.03YY353 pKa = 11.09LSKK356 pKa = 10.76VGRR359 pKa = 11.84DD360 pKa = 3.25LTGFPSADD368 pKa = 3.25AATLPGVRR376 pKa = 11.84AADD379 pKa = 3.41GFLPAEE385 pKa = 4.47HH386 pKa = 7.0EE387 pKa = 4.37WGSMYY392 pKa = 10.31YY393 pKa = 9.68IPNYY397 pKa = 9.42TVTNGDD403 pKa = 4.11SIVADD408 pKa = 3.35SFLYY412 pKa = 10.25TGATVNADD420 pKa = 3.5TSVTEE425 pKa = 3.9AVVQVRR431 pKa = 11.84LYY433 pKa = 10.97KK434 pKa = 10.95FNDD437 pKa = 3.51DD438 pKa = 4.51DD439 pKa = 6.38GSGFWTAQPTGPSDD453 pKa = 3.73PEE455 pKa = 4.15LPIQALGVDD464 pKa = 4.04TFEE467 pKa = 6.34LDD469 pKa = 2.85LSLAFRR475 pKa = 11.84QRR477 pKa = 11.84AVALSNLSGGTGAVLQPNSFYY498 pKa = 11.19VATVLCSDD506 pKa = 4.58QINGLRR512 pKa = 11.84NADD515 pKa = 3.15NTTRR519 pKa = 11.84HH520 pKa = 5.39IFIGNSEE527 pKa = 4.45DD528 pKa = 4.74KK529 pKa = 11.19YY530 pKa = 11.58DD531 pKa = 3.73FTIDD535 pKa = 3.38SLPNYY540 pKa = 7.64FIPASIVRR548 pKa = 11.84SAYY551 pKa = 9.3IDD553 pKa = 3.33PTGTVTGLQDD563 pKa = 3.73NQWYY567 pKa = 8.39GAGYY571 pKa = 9.98EE572 pKa = 4.15SGIVPSIGLTLTDD585 pKa = 5.55AITINVTANEE595 pKa = 3.99AVTTEE600 pKa = 4.41FNIFPNPATEE610 pKa = 4.05QISAKK615 pKa = 10.52VEE617 pKa = 3.95LTEE620 pKa = 4.02MNSNVEE626 pKa = 4.13YY627 pKa = 10.33IITDD631 pKa = 3.08ATGRR635 pKa = 11.84VVIKK639 pKa = 8.8EE640 pKa = 4.01QKK642 pKa = 10.67ADD644 pKa = 3.67LQSDD648 pKa = 4.2VVTYY652 pKa = 10.52DD653 pKa = 3.19VSQLPAGVYY662 pKa = 9.43FFTVRR667 pKa = 11.84TAQGASSQKK676 pKa = 9.9FVKK679 pKa = 10.06QQ680 pKa = 3.29
Molecular weight: 72.49 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|H6L936|H6L936_SAPGL Uncharacterized protein OS=Saprospira grandis (strain Lewin) OX=984262 GN=SGRA_0433 PE=4 SV=1
MM1 pKa = 7.48SGSKK5 pKa = 10.0LVRR8 pKa = 11.84SGGKK12 pKa = 9.25LVRR15 pKa = 11.84SGGKK19 pKa = 9.25LVRR22 pKa = 11.84SGGKK26 pKa = 9.25LVRR29 pKa = 11.84SGGKK33 pKa = 9.27LVRR36 pKa = 11.84SGSKK40 pKa = 9.93LVRR43 pKa = 11.84FGGKK47 pKa = 9.63LVMSGGKK54 pKa = 9.25LVRR57 pKa = 11.84SGGKK61 pKa = 9.27LVRR64 pKa = 11.84SGSKK68 pKa = 10.53LVMSGSKK75 pKa = 10.0LVRR78 pKa = 11.84SGGKK82 pKa = 9.83LVMSGSKK89 pKa = 10.0LVRR92 pKa = 11.84SGGKK96 pKa = 9.19LVRR99 pKa = 11.84FGGKK103 pKa = 9.36LVMPIVAA110 pKa = 4.7
MM1 pKa = 7.48SGSKK5 pKa = 10.0LVRR8 pKa = 11.84SGGKK12 pKa = 9.25LVRR15 pKa = 11.84SGGKK19 pKa = 9.25LVRR22 pKa = 11.84SGGKK26 pKa = 9.25LVRR29 pKa = 11.84SGGKK33 pKa = 9.27LVRR36 pKa = 11.84SGSKK40 pKa = 9.93LVRR43 pKa = 11.84FGGKK47 pKa = 9.63LVMSGGKK54 pKa = 9.25LVRR57 pKa = 11.84SGGKK61 pKa = 9.27LVRR64 pKa = 11.84SGSKK68 pKa = 10.53LVMSGSKK75 pKa = 10.0LVRR78 pKa = 11.84SGGKK82 pKa = 9.83LVMSGSKK89 pKa = 10.0LVRR92 pKa = 11.84SGGKK96 pKa = 9.19LVRR99 pKa = 11.84FGGKK103 pKa = 9.36LVMPIVAA110 pKa = 4.7
Molecular weight: 11.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1241634 |
30 |
3619 |
294.4 |
33.16 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.746 ± 0.054 | 1.016 ± 0.021 |
4.9 ± 0.035 | 6.991 ± 0.055 |
4.8 ± 0.033 | 6.647 ± 0.049 |
1.693 ± 0.021 | 5.636 ± 0.033 |
6.348 ± 0.071 | 11.369 ± 0.077 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.215 ± 0.022 | 4.263 ± 0.038 |
4.27 ± 0.029 | 5.409 ± 0.042 |
4.744 ± 0.042 | 6.46 ± 0.045 |
4.051 ± 0.071 | 5.033 ± 0.04 |
1.389 ± 0.018 | 4.018 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
