
Rhodococcus kyotonensis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Corynebacteriales; Nocardiaceae; Rhodococcus
Average proteome isoelectric point is 5.82
Get precalculated fractions of proteins
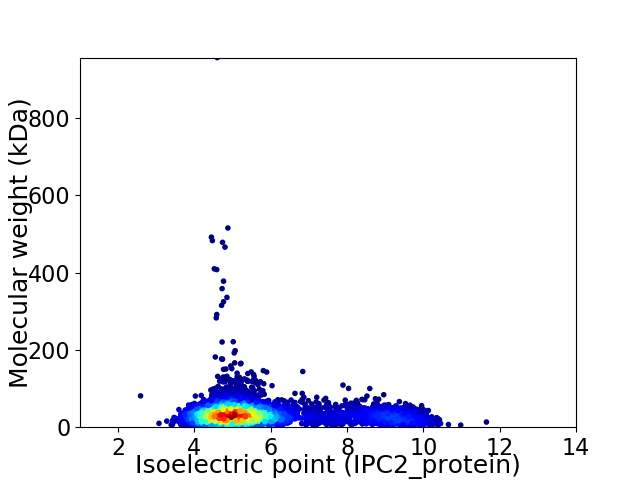
Virtual 2D-PAGE plot for 4877 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A177YKY3|A0A177YKY3_9NOCA Glycosyl transferase family 1 OS=Rhodococcus kyotonensis OX=398843 GN=A3K89_17900 PE=4 SV=1
MM1 pKa = 7.62GGGAVKK7 pKa = 10.44KK8 pKa = 10.34LLALAVSAVSIAGLTACAQSNDD30 pKa = 2.97SDD32 pKa = 4.18TIRR35 pKa = 11.84FALDD39 pKa = 2.82WTPNTNHH46 pKa = 6.56TGLYY50 pKa = 9.41VALQEE55 pKa = 5.7GYY57 pKa = 10.07FADD60 pKa = 3.99AGLDD64 pKa = 3.87VEE66 pKa = 4.45VLPYY70 pKa = 11.26NNTSPDD76 pKa = 3.51TLVDD80 pKa = 3.35SGNAEE85 pKa = 4.27FGISTQSSTAFAKK98 pKa = 10.68AAGAQIVSVLAPLQHH113 pKa = 6.72WATGIGVRR121 pKa = 11.84ADD123 pKa = 3.87RR124 pKa = 11.84DD125 pKa = 4.34DD126 pKa = 3.9IPSPRR131 pKa = 11.84ALDD134 pKa = 3.61GKK136 pKa = 8.94TYY138 pKa = 10.98AGFGDD143 pKa = 3.79TGEE146 pKa = 4.2QATLRR151 pKa = 11.84QVVRR155 pKa = 11.84NDD157 pKa = 3.22GGAGEE162 pKa = 4.61YY163 pKa = 7.46EE164 pKa = 4.39TVVLGTSAYY173 pKa = 8.24EE174 pKa = 3.84AVYY177 pKa = 10.71SGTADD182 pKa = 3.42FTVSYY187 pKa = 8.35FAWEE191 pKa = 4.79GIEE194 pKa = 4.44AEE196 pKa = 4.44HH197 pKa = 6.86SGTPMKK203 pKa = 10.96YY204 pKa = 10.06FDD206 pKa = 3.55YY207 pKa = 10.45TDD209 pKa = 3.24YY210 pKa = 11.5GFPDD214 pKa = 4.17AYY216 pKa = 10.58AITISGNEE224 pKa = 3.54NWLADD229 pKa = 3.95NPEE232 pKa = 4.29DD233 pKa = 3.53ATKK236 pKa = 10.19FVQALQRR243 pKa = 11.84GYY245 pKa = 10.72QLAADD250 pKa = 4.46DD251 pKa = 4.26PHH253 pKa = 7.22RR254 pKa = 11.84AAQDD258 pKa = 3.61VIDD261 pKa = 4.82ANPGAFTDD269 pKa = 3.73EE270 pKa = 4.05NLVFEE275 pKa = 4.59SQQMLADD282 pKa = 4.29SYY284 pKa = 11.21MKK286 pKa = 10.48DD287 pKa = 2.92ASGTVGTQNLDD298 pKa = 2.45KK299 pKa = 10.12WAAYY303 pKa = 10.67SKK305 pKa = 10.4FLYY308 pKa = 10.78DD309 pKa = 3.47NGVLVDD315 pKa = 3.96EE316 pKa = 5.25SGAPLTSEE324 pKa = 5.03PDD326 pKa = 2.7WSTYY330 pKa = 8.29FTDD333 pKa = 6.18DD334 pKa = 3.65YY335 pKa = 11.64LADD338 pKa = 3.51QQ339 pKa = 4.39
MM1 pKa = 7.62GGGAVKK7 pKa = 10.44KK8 pKa = 10.34LLALAVSAVSIAGLTACAQSNDD30 pKa = 2.97SDD32 pKa = 4.18TIRR35 pKa = 11.84FALDD39 pKa = 2.82WTPNTNHH46 pKa = 6.56TGLYY50 pKa = 9.41VALQEE55 pKa = 5.7GYY57 pKa = 10.07FADD60 pKa = 3.99AGLDD64 pKa = 3.87VEE66 pKa = 4.45VLPYY70 pKa = 11.26NNTSPDD76 pKa = 3.51TLVDD80 pKa = 3.35SGNAEE85 pKa = 4.27FGISTQSSTAFAKK98 pKa = 10.68AAGAQIVSVLAPLQHH113 pKa = 6.72WATGIGVRR121 pKa = 11.84ADD123 pKa = 3.87RR124 pKa = 11.84DD125 pKa = 4.34DD126 pKa = 3.9IPSPRR131 pKa = 11.84ALDD134 pKa = 3.61GKK136 pKa = 8.94TYY138 pKa = 10.98AGFGDD143 pKa = 3.79TGEE146 pKa = 4.2QATLRR151 pKa = 11.84QVVRR155 pKa = 11.84NDD157 pKa = 3.22GGAGEE162 pKa = 4.61YY163 pKa = 7.46EE164 pKa = 4.39TVVLGTSAYY173 pKa = 8.24EE174 pKa = 3.84AVYY177 pKa = 10.71SGTADD182 pKa = 3.42FTVSYY187 pKa = 8.35FAWEE191 pKa = 4.79GIEE194 pKa = 4.44AEE196 pKa = 4.44HH197 pKa = 6.86SGTPMKK203 pKa = 10.96YY204 pKa = 10.06FDD206 pKa = 3.55YY207 pKa = 10.45TDD209 pKa = 3.24YY210 pKa = 11.5GFPDD214 pKa = 4.17AYY216 pKa = 10.58AITISGNEE224 pKa = 3.54NWLADD229 pKa = 3.95NPEE232 pKa = 4.29DD233 pKa = 3.53ATKK236 pKa = 10.19FVQALQRR243 pKa = 11.84GYY245 pKa = 10.72QLAADD250 pKa = 4.46DD251 pKa = 4.26PHH253 pKa = 7.22RR254 pKa = 11.84AAQDD258 pKa = 3.61VIDD261 pKa = 4.82ANPGAFTDD269 pKa = 3.73EE270 pKa = 4.05NLVFEE275 pKa = 4.59SQQMLADD282 pKa = 4.29SYY284 pKa = 11.21MKK286 pKa = 10.48DD287 pKa = 2.92ASGTVGTQNLDD298 pKa = 2.45KK299 pKa = 10.12WAAYY303 pKa = 10.67SKK305 pKa = 10.4FLYY308 pKa = 10.78DD309 pKa = 3.47NGVLVDD315 pKa = 3.96EE316 pKa = 5.25SGAPLTSEE324 pKa = 5.03PDD326 pKa = 2.7WSTYY330 pKa = 8.29FTDD333 pKa = 6.18DD334 pKa = 3.65YY335 pKa = 11.64LADD338 pKa = 3.51QQ339 pKa = 4.39
Molecular weight: 36.27 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A177Y9F6|A0A177Y9F6_9NOCA Uncharacterized protein OS=Rhodococcus kyotonensis OX=398843 GN=A3K89_10830 PE=4 SV=1
MM1 pKa = 7.69AKK3 pKa = 10.06GKK5 pKa = 8.69RR6 pKa = 11.84TFQPNNRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84ARR17 pKa = 11.84VHH19 pKa = 5.99GFRR22 pKa = 11.84LRR24 pKa = 11.84MRR26 pKa = 11.84TRR28 pKa = 11.84AGRR31 pKa = 11.84AIVSARR37 pKa = 11.84RR38 pKa = 11.84GKK40 pKa = 10.01GRR42 pKa = 11.84KK43 pKa = 8.43EE44 pKa = 3.47LTAA47 pKa = 4.94
MM1 pKa = 7.69AKK3 pKa = 10.06GKK5 pKa = 8.69RR6 pKa = 11.84TFQPNNRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84ARR17 pKa = 11.84VHH19 pKa = 5.99GFRR22 pKa = 11.84LRR24 pKa = 11.84MRR26 pKa = 11.84TRR28 pKa = 11.84AGRR31 pKa = 11.84AIVSARR37 pKa = 11.84RR38 pKa = 11.84GKK40 pKa = 10.01GRR42 pKa = 11.84KK43 pKa = 8.43EE44 pKa = 3.47LTAA47 pKa = 4.94
Molecular weight: 5.48 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1626480 |
33 |
8883 |
333.5 |
35.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.5 ± 0.046 | 0.676 ± 0.01 |
6.717 ± 0.033 | 5.436 ± 0.031 |
3.112 ± 0.021 | 8.72 ± 0.03 |
2.104 ± 0.016 | 4.354 ± 0.024 |
2.319 ± 0.028 | 9.753 ± 0.03 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.968 ± 0.015 | 2.129 ± 0.021 |
5.261 ± 0.025 | 2.728 ± 0.018 |
6.925 ± 0.032 | 6.205 ± 0.021 |
6.354 ± 0.023 | 9.301 ± 0.044 |
1.391 ± 0.014 | 2.044 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
