
Halomonas sp. ND22Bw
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Oceanospirillales; Halomonadaceae; Halomonas; unclassified Halomonas
Average proteome isoelectric point is 6.12
Get precalculated fractions of proteins
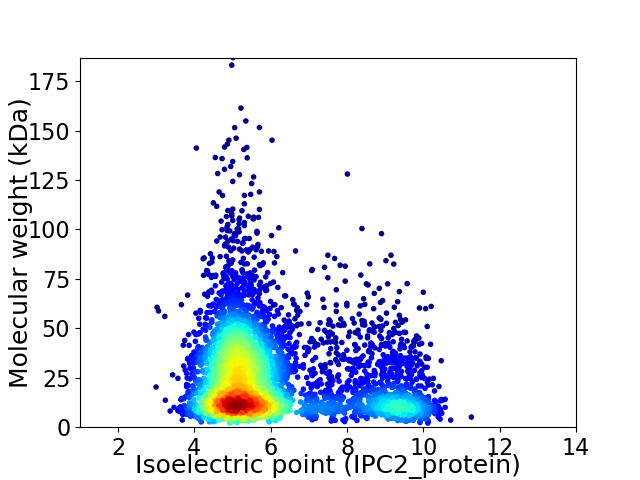
Virtual 2D-PAGE plot for 4859 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
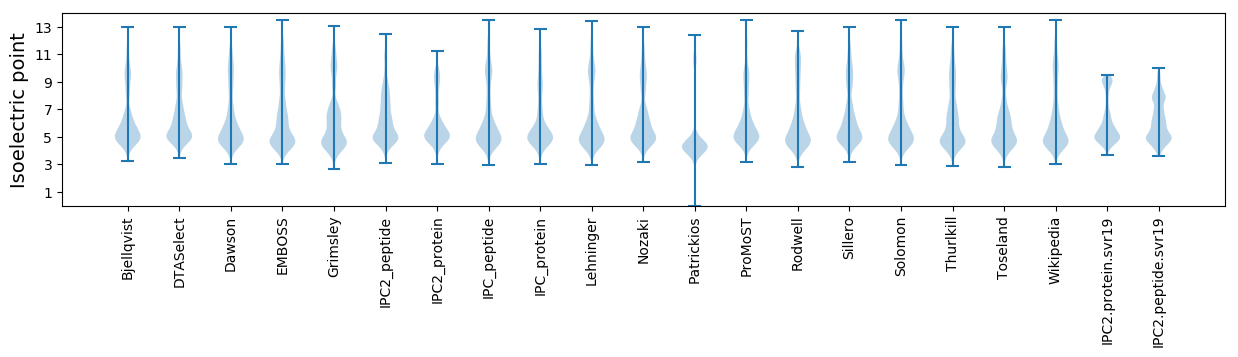
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2P7P843|A0A2P7P843_9GAMM FUSC family protein OS=Halomonas sp. ND22Bw OX=2054178 GN=CVH10_09575 PE=4 SV=1
MM1 pKa = 7.71KK2 pKa = 9.06KK3 pKa = 8.27TLLATAIAGAMAASGAQAATVYY25 pKa = 10.61NQDD28 pKa = 3.01GTKK31 pKa = 10.46LDD33 pKa = 3.42IYY35 pKa = 11.34GNLQIAYY42 pKa = 9.67RR43 pKa = 11.84SIEE46 pKa = 4.09TADD49 pKa = 3.56EE50 pKa = 4.21STGDD54 pKa = 3.39VEE56 pKa = 5.07SRR58 pKa = 11.84DD59 pKa = 4.28DD60 pKa = 3.61IFDD63 pKa = 3.86NGSTLGVAGEE73 pKa = 3.99HH74 pKa = 6.35VIYY77 pKa = 10.78DD78 pKa = 3.7GLTGYY83 pKa = 10.22FKK85 pKa = 11.68YY86 pKa = 10.38EE87 pKa = 3.83FEE89 pKa = 4.57GNADD93 pKa = 3.61EE94 pKa = 6.21AKK96 pKa = 10.79DD97 pKa = 3.61QGGLNSGDD105 pKa = 3.2QAYY108 pKa = 10.31FGLKK112 pKa = 10.51GNFGDD117 pKa = 4.77ARR119 pKa = 11.84LGSWDD124 pKa = 4.23PLIDD128 pKa = 5.15DD129 pKa = 5.77WIQDD133 pKa = 3.91PITNNEE139 pKa = 3.92YY140 pKa = 10.91FDD142 pKa = 4.42VSDD145 pKa = 3.45SSSRR149 pKa = 11.84IVGVEE154 pKa = 3.93DD155 pKa = 3.26SRR157 pKa = 11.84EE158 pKa = 4.18GNKK161 pKa = 9.39LQYY164 pKa = 9.29MSPSFSGFQFAVGTQYY180 pKa = 11.15EE181 pKa = 4.3GDD183 pKa = 3.77AEE185 pKa = 4.14YY186 pKa = 10.77DD187 pKa = 3.38ASNTASISGTNGANLTVSEE206 pKa = 4.76KK207 pKa = 10.25RR208 pKa = 11.84DD209 pKa = 3.53SNASFFGGVKK219 pKa = 8.36YY220 pKa = 8.62TAGAFSIAAVYY231 pKa = 10.89DD232 pKa = 3.62NLDD235 pKa = 4.02NNDD238 pKa = 2.77GRR240 pKa = 11.84YY241 pKa = 10.49DD242 pKa = 3.76GVDD245 pKa = 2.93GDD247 pKa = 4.77GNVISGDD254 pKa = 3.38FDD256 pKa = 4.35AGEE259 pKa = 4.18QYY261 pKa = 11.05GITAQYY267 pKa = 8.75TWDD270 pKa = 3.81TLRR273 pKa = 11.84VAAKK277 pKa = 9.71VEE279 pKa = 4.2RR280 pKa = 11.84FKK282 pKa = 11.48SDD284 pKa = 2.86NDD286 pKa = 3.48FVADD290 pKa = 3.72TNFYY294 pKa = 11.11GLGARR299 pKa = 11.84YY300 pKa = 9.66GYY302 pKa = 11.31GNGDD306 pKa = 2.74IYY308 pKa = 11.26GAYY311 pKa = 9.92QYY313 pKa = 11.46VDD315 pKa = 3.28VGGGDD320 pKa = 4.89LIDD323 pKa = 4.14TADD326 pKa = 4.02DD327 pKa = 4.17ALTSGDD333 pKa = 3.48WPNEE337 pKa = 4.05RR338 pKa = 11.84EE339 pKa = 4.02DD340 pKa = 3.58EE341 pKa = 4.41SYY343 pKa = 11.62NEE345 pKa = 3.86IMLGATYY352 pKa = 10.55NVSDD356 pKa = 3.38AMYY359 pKa = 9.4TFVEE363 pKa = 4.31AAWYY367 pKa = 9.89DD368 pKa = 3.55RR369 pKa = 11.84DD370 pKa = 3.69EE371 pKa = 4.55DD372 pKa = 3.87VGDD375 pKa = 4.18GVAVGAVYY383 pKa = 10.44LFF385 pKa = 4.21
MM1 pKa = 7.71KK2 pKa = 9.06KK3 pKa = 8.27TLLATAIAGAMAASGAQAATVYY25 pKa = 10.61NQDD28 pKa = 3.01GTKK31 pKa = 10.46LDD33 pKa = 3.42IYY35 pKa = 11.34GNLQIAYY42 pKa = 9.67RR43 pKa = 11.84SIEE46 pKa = 4.09TADD49 pKa = 3.56EE50 pKa = 4.21STGDD54 pKa = 3.39VEE56 pKa = 5.07SRR58 pKa = 11.84DD59 pKa = 4.28DD60 pKa = 3.61IFDD63 pKa = 3.86NGSTLGVAGEE73 pKa = 3.99HH74 pKa = 6.35VIYY77 pKa = 10.78DD78 pKa = 3.7GLTGYY83 pKa = 10.22FKK85 pKa = 11.68YY86 pKa = 10.38EE87 pKa = 3.83FEE89 pKa = 4.57GNADD93 pKa = 3.61EE94 pKa = 6.21AKK96 pKa = 10.79DD97 pKa = 3.61QGGLNSGDD105 pKa = 3.2QAYY108 pKa = 10.31FGLKK112 pKa = 10.51GNFGDD117 pKa = 4.77ARR119 pKa = 11.84LGSWDD124 pKa = 4.23PLIDD128 pKa = 5.15DD129 pKa = 5.77WIQDD133 pKa = 3.91PITNNEE139 pKa = 3.92YY140 pKa = 10.91FDD142 pKa = 4.42VSDD145 pKa = 3.45SSSRR149 pKa = 11.84IVGVEE154 pKa = 3.93DD155 pKa = 3.26SRR157 pKa = 11.84EE158 pKa = 4.18GNKK161 pKa = 9.39LQYY164 pKa = 9.29MSPSFSGFQFAVGTQYY180 pKa = 11.15EE181 pKa = 4.3GDD183 pKa = 3.77AEE185 pKa = 4.14YY186 pKa = 10.77DD187 pKa = 3.38ASNTASISGTNGANLTVSEE206 pKa = 4.76KK207 pKa = 10.25RR208 pKa = 11.84DD209 pKa = 3.53SNASFFGGVKK219 pKa = 8.36YY220 pKa = 8.62TAGAFSIAAVYY231 pKa = 10.89DD232 pKa = 3.62NLDD235 pKa = 4.02NNDD238 pKa = 2.77GRR240 pKa = 11.84YY241 pKa = 10.49DD242 pKa = 3.76GVDD245 pKa = 2.93GDD247 pKa = 4.77GNVISGDD254 pKa = 3.38FDD256 pKa = 4.35AGEE259 pKa = 4.18QYY261 pKa = 11.05GITAQYY267 pKa = 8.75TWDD270 pKa = 3.81TLRR273 pKa = 11.84VAAKK277 pKa = 9.71VEE279 pKa = 4.2RR280 pKa = 11.84FKK282 pKa = 11.48SDD284 pKa = 2.86NDD286 pKa = 3.48FVADD290 pKa = 3.72TNFYY294 pKa = 11.11GLGARR299 pKa = 11.84YY300 pKa = 9.66GYY302 pKa = 11.31GNGDD306 pKa = 2.74IYY308 pKa = 11.26GAYY311 pKa = 9.92QYY313 pKa = 11.46VDD315 pKa = 3.28VGGGDD320 pKa = 4.89LIDD323 pKa = 4.14TADD326 pKa = 4.02DD327 pKa = 4.17ALTSGDD333 pKa = 3.48WPNEE337 pKa = 4.05RR338 pKa = 11.84EE339 pKa = 4.02DD340 pKa = 3.58EE341 pKa = 4.41SYY343 pKa = 11.62NEE345 pKa = 3.86IMLGATYY352 pKa = 10.55NVSDD356 pKa = 3.38AMYY359 pKa = 9.4TFVEE363 pKa = 4.31AAWYY367 pKa = 9.89DD368 pKa = 3.55RR369 pKa = 11.84DD370 pKa = 3.69EE371 pKa = 4.55DD372 pKa = 3.87VGDD375 pKa = 4.18GVAVGAVYY383 pKa = 10.44LFF385 pKa = 4.21
Molecular weight: 41.49 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2P7PAJ7|A0A2P7PAJ7_9GAMM ATP-dependent DNA helicase RecG OS=Halomonas sp. ND22Bw OX=2054178 GN=recG PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.42RR12 pKa = 11.84KK13 pKa = 9.04RR14 pKa = 11.84AHH16 pKa = 6.12GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.3NGRR28 pKa = 11.84AVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.93GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.42RR12 pKa = 11.84KK13 pKa = 9.04RR14 pKa = 11.84AHH16 pKa = 6.12GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.3NGRR28 pKa = 11.84AVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.93GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1204888 |
20 |
1644 |
248.0 |
27.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.132 ± 0.052 | 0.877 ± 0.012 |
6.1 ± 0.038 | 6.671 ± 0.046 |
3.337 ± 0.026 | 8.645 ± 0.037 |
2.398 ± 0.019 | 4.231 ± 0.032 |
2.342 ± 0.034 | 11.457 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.42 ± 0.019 | 2.226 ± 0.02 |
5.09 ± 0.027 | 3.465 ± 0.024 |
7.797 ± 0.051 | 5.022 ± 0.025 |
4.883 ± 0.025 | 7.219 ± 0.035 |
1.458 ± 0.019 | 2.229 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
