
Marisediminitalea aggregata
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Alteromonadaceae; Marisediminitalea
Average proteome isoelectric point is 5.93
Get precalculated fractions of proteins
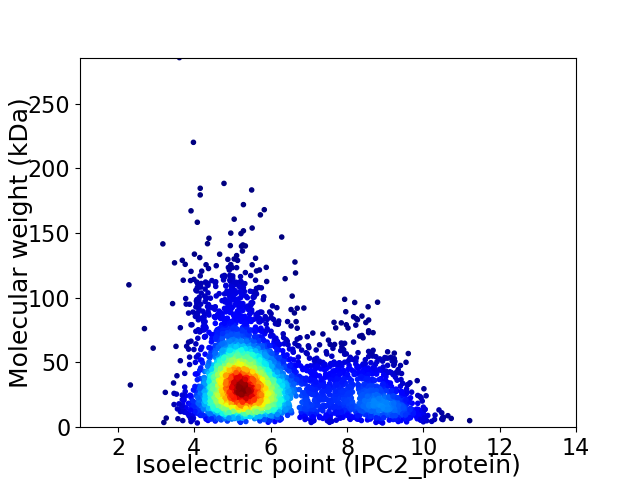
Virtual 2D-PAGE plot for 4313 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1M5HVM5|A0A1M5HVM5_9ALTE Transporter SSS family OS=Marisediminitalea aggregata OX=634436 GN=SAMN05216361_1638 PE=3 SV=1
MM1 pKa = 7.46KK2 pKa = 9.94IKK4 pKa = 10.79YY5 pKa = 7.57PALISAMWASQVFATDD21 pKa = 2.96IFINEE26 pKa = 3.87LHH28 pKa = 6.48YY29 pKa = 11.25DD30 pKa = 3.51NSGGDD35 pKa = 3.0VGEE38 pKa = 4.27FVEE41 pKa = 5.26VVAPAGTDD49 pKa = 3.21LTGYY53 pKa = 10.34KK54 pKa = 10.22VLFYY58 pKa = 10.74NGSNGTQYY66 pKa = 11.36NSRR69 pKa = 11.84EE70 pKa = 4.16LVGLVADD77 pKa = 4.29MGNGFGTYY85 pKa = 10.08AIDD88 pKa = 3.96VSGIQNGAPDD98 pKa = 4.28GVALVDD104 pKa = 3.72ASSNVIQFLSYY115 pKa = 10.33EE116 pKa = 4.27GSFTATAGVLAGLTSVDD133 pKa = 3.02IGVSQSSSTPAGASLQLTGTGAQYY157 pKa = 11.48SDD159 pKa = 3.99FTWQAFTTATKK170 pKa = 10.6DD171 pKa = 3.37AVNEE175 pKa = 4.23GQTFTGNEE183 pKa = 3.63VVFFNEE189 pKa = 3.07IHH191 pKa = 6.42YY192 pKa = 10.81DD193 pKa = 3.52NAGGDD198 pKa = 3.25VGEE201 pKa = 4.96FIEE204 pKa = 4.5IAGTAGLDD212 pKa = 3.51LTGMTIEE219 pKa = 5.47LYY221 pKa = 10.4NGSNGTVYY229 pKa = 9.63NTIVLSGALPDD240 pKa = 3.85NGNGFGTIAYY250 pKa = 8.48YY251 pKa = 10.02PSSIQNGAPDD261 pKa = 3.75GMALVSATGEE271 pKa = 4.19VIQFLSYY278 pKa = 10.61EE279 pKa = 4.4GVVSATAGAAVGTDD293 pKa = 3.24STDD296 pKa = 2.67IGIAEE301 pKa = 4.39TSSTAIGQSLQLSGTGTRR319 pKa = 11.84YY320 pKa = 10.44SDD322 pKa = 3.65FTWIAGGEE330 pKa = 4.4TPDD333 pKa = 3.81NTNASQTFEE342 pKa = 4.18GVGGGTPEE350 pKa = 3.97EE351 pKa = 4.4PEE353 pKa = 4.26EE354 pKa = 4.44PEE356 pKa = 4.22EE357 pKa = 4.08PTPGTGDD364 pKa = 3.4VAKK367 pKa = 10.93LFINEE372 pKa = 3.94LHH374 pKa = 6.5YY375 pKa = 11.52DD376 pKa = 3.66NASSDD381 pKa = 3.23VGEE384 pKa = 4.37FVEE387 pKa = 4.41IAGPAGTDD395 pKa = 3.27LTGVSIEE402 pKa = 4.64LYY404 pKa = 9.51NGNGGGVYY412 pKa = 8.68KK413 pKa = 10.27TVALSGTIPEE423 pKa = 4.17QAGGFGTVAEE433 pKa = 4.82YY434 pKa = 10.41ISGIQNGAPDD444 pKa = 4.66GVALSYY450 pKa = 10.93NGEE453 pKa = 4.39LIDD456 pKa = 4.44FISYY460 pKa = 10.18EE461 pKa = 4.32GEE463 pKa = 3.82LTATGGSASGVTATDD478 pKa = 2.63IGVAEE483 pKa = 4.38TGSTLSTEE491 pKa = 4.58SVQLTGTGSEE501 pKa = 4.21VCHH504 pKa = 6.31FSWSGPATSSPGEE517 pKa = 3.97INAGQTFDD525 pKa = 3.67ASVGATCGGNDD536 pKa = 4.05GGDD539 pKa = 3.29GGTGGEE545 pKa = 4.19VTLVKK550 pKa = 10.18ISEE553 pKa = 4.38VQGSGSSSPLINAQVAIEE571 pKa = 4.64AIVVADD577 pKa = 3.97HH578 pKa = 6.24QSGSGTDD585 pKa = 3.18GDD587 pKa = 3.86LRR589 pKa = 11.84GFFVQEE595 pKa = 3.56EE596 pKa = 4.23DD597 pKa = 3.65ADD599 pKa = 4.06ADD601 pKa = 4.07ADD603 pKa = 3.95SSTSEE608 pKa = 3.9GLFVFDD614 pKa = 4.65GSSPAVDD621 pKa = 3.33VAVGDD626 pKa = 3.67KK627 pKa = 10.6VYY629 pKa = 11.07LVGTVDD635 pKa = 3.99EE636 pKa = 4.82YY637 pKa = 11.51FGDD640 pKa = 3.78TQLDD644 pKa = 3.95VQSGSVQVVSSGNALPSPAIVTLPLATTTNSGGEE678 pKa = 4.12AIADD682 pKa = 3.98LEE684 pKa = 4.61AYY686 pKa = 10.0EE687 pKa = 4.61GMQVMFAQTLAVTEE701 pKa = 4.48LFNLDD706 pKa = 3.23RR707 pKa = 11.84FGEE710 pKa = 4.15ILLSSEE716 pKa = 3.9GRR718 pKa = 11.84QVQYY722 pKa = 10.61TEE724 pKa = 3.61IMTPSVEE731 pKa = 4.28GYY733 pKa = 9.5AAYY736 pKa = 10.23VASAAANRR744 pKa = 11.84LMLDD748 pKa = 3.95DD749 pKa = 5.41GFSIQNPSPIRR760 pKa = 11.84FPAPGLSTANTVRR773 pKa = 11.84MSDD776 pKa = 3.18EE777 pKa = 4.18VTNLTGVISYY787 pKa = 10.3RR788 pKa = 11.84RR789 pKa = 11.84GSGGSGDD796 pKa = 3.3EE797 pKa = 4.64MYY799 pKa = 11.26RR800 pKa = 11.84LMPTVEE806 pKa = 4.2PVFTRR811 pKa = 11.84PEE813 pKa = 4.0SRR815 pKa = 11.84KK816 pKa = 9.97DD817 pKa = 3.5RR818 pKa = 11.84PDD820 pKa = 3.13VGGSIQVASFNLLNFFNTFGNSCYY844 pKa = 10.41LGGGTGDD851 pKa = 4.61CRR853 pKa = 11.84GADD856 pKa = 3.3NTEE859 pKa = 3.96EE860 pKa = 4.72YY861 pKa = 10.73DD862 pKa = 3.59RR863 pKa = 11.84QLQKK867 pKa = 9.98TVTAITALDD876 pKa = 3.83ADD878 pKa = 3.99VLGVIEE884 pKa = 4.5IEE886 pKa = 3.86NDD888 pKa = 2.9IVEE891 pKa = 4.61GQNSAMASLVNALNQAGSACSDD913 pKa = 3.55YY914 pKa = 11.19QYY916 pKa = 11.28VDD918 pKa = 3.16PQQRR922 pKa = 11.84VGTDD926 pKa = 3.95AIAVGFLYY934 pKa = 10.84CADD937 pKa = 3.9TVEE940 pKa = 4.46VTAGSSIAILDD951 pKa = 3.99DD952 pKa = 3.73SQLAGLGLSDD962 pKa = 4.09YY963 pKa = 11.42APVFDD968 pKa = 5.04GSSTSRR974 pKa = 11.84PSLTVSFTEE983 pKa = 4.17KK984 pKa = 9.81ATSEE988 pKa = 4.27VFTVSVNHH996 pKa = 6.4FKK998 pKa = 11.19SKK1000 pKa = 10.51GGSGSGGNADD1010 pKa = 4.34ANDD1013 pKa = 4.2GAGNWNEE1020 pKa = 3.8RR1021 pKa = 11.84RR1022 pKa = 11.84TQAAEE1027 pKa = 4.13ALHH1030 pKa = 6.59KK1031 pKa = 10.11WLATYY1036 pKa = 7.59PTGVSDD1042 pKa = 6.11DD1043 pKa = 5.08DD1044 pKa = 3.59ILIIGDD1050 pKa = 4.08LNAYY1054 pKa = 10.38SMEE1057 pKa = 4.26DD1058 pKa = 3.69PIVEE1062 pKa = 4.23LTSNGYY1068 pKa = 8.01TNVTGGEE1075 pKa = 3.67AHH1077 pKa = 6.19YY1078 pKa = 10.81SYY1080 pKa = 11.49VFDD1083 pKa = 4.39GFAGNLDD1090 pKa = 3.84HH1091 pKa = 7.1GLASASLMDD1100 pKa = 4.6QITGATSWNINSDD1113 pKa = 3.36EE1114 pKa = 4.75ADD1116 pKa = 3.46ALDD1119 pKa = 3.78YY1120 pKa = 10.35NTDD1123 pKa = 3.76FGRR1126 pKa = 11.84PTDD1129 pKa = 3.76IFDD1132 pKa = 3.45GTTPYY1137 pKa = 9.19RR1138 pKa = 11.84TSDD1141 pKa = 3.79HH1142 pKa = 7.29DD1143 pKa = 3.93PLLIGLSLSSVLKK1156 pKa = 10.79GDD1158 pKa = 3.97LDD1160 pKa = 4.21GDD1162 pKa = 4.04GDD1164 pKa = 4.11IDD1166 pKa = 4.79RR1167 pKa = 11.84MDD1169 pKa = 5.65FILIMRR1175 pKa = 11.84AILTRR1180 pKa = 11.84KK1181 pKa = 9.6PFDD1184 pKa = 3.48AANDD1188 pKa = 3.65INGDD1192 pKa = 3.78GVVNIVDD1199 pKa = 3.65GLLLRR1204 pKa = 11.84RR1205 pKa = 11.84SCTRR1209 pKa = 11.84ARR1211 pKa = 11.84CAVRR1215 pKa = 4.07
MM1 pKa = 7.46KK2 pKa = 9.94IKK4 pKa = 10.79YY5 pKa = 7.57PALISAMWASQVFATDD21 pKa = 2.96IFINEE26 pKa = 3.87LHH28 pKa = 6.48YY29 pKa = 11.25DD30 pKa = 3.51NSGGDD35 pKa = 3.0VGEE38 pKa = 4.27FVEE41 pKa = 5.26VVAPAGTDD49 pKa = 3.21LTGYY53 pKa = 10.34KK54 pKa = 10.22VLFYY58 pKa = 10.74NGSNGTQYY66 pKa = 11.36NSRR69 pKa = 11.84EE70 pKa = 4.16LVGLVADD77 pKa = 4.29MGNGFGTYY85 pKa = 10.08AIDD88 pKa = 3.96VSGIQNGAPDD98 pKa = 4.28GVALVDD104 pKa = 3.72ASSNVIQFLSYY115 pKa = 10.33EE116 pKa = 4.27GSFTATAGVLAGLTSVDD133 pKa = 3.02IGVSQSSSTPAGASLQLTGTGAQYY157 pKa = 11.48SDD159 pKa = 3.99FTWQAFTTATKK170 pKa = 10.6DD171 pKa = 3.37AVNEE175 pKa = 4.23GQTFTGNEE183 pKa = 3.63VVFFNEE189 pKa = 3.07IHH191 pKa = 6.42YY192 pKa = 10.81DD193 pKa = 3.52NAGGDD198 pKa = 3.25VGEE201 pKa = 4.96FIEE204 pKa = 4.5IAGTAGLDD212 pKa = 3.51LTGMTIEE219 pKa = 5.47LYY221 pKa = 10.4NGSNGTVYY229 pKa = 9.63NTIVLSGALPDD240 pKa = 3.85NGNGFGTIAYY250 pKa = 8.48YY251 pKa = 10.02PSSIQNGAPDD261 pKa = 3.75GMALVSATGEE271 pKa = 4.19VIQFLSYY278 pKa = 10.61EE279 pKa = 4.4GVVSATAGAAVGTDD293 pKa = 3.24STDD296 pKa = 2.67IGIAEE301 pKa = 4.39TSSTAIGQSLQLSGTGTRR319 pKa = 11.84YY320 pKa = 10.44SDD322 pKa = 3.65FTWIAGGEE330 pKa = 4.4TPDD333 pKa = 3.81NTNASQTFEE342 pKa = 4.18GVGGGTPEE350 pKa = 3.97EE351 pKa = 4.4PEE353 pKa = 4.26EE354 pKa = 4.44PEE356 pKa = 4.22EE357 pKa = 4.08PTPGTGDD364 pKa = 3.4VAKK367 pKa = 10.93LFINEE372 pKa = 3.94LHH374 pKa = 6.5YY375 pKa = 11.52DD376 pKa = 3.66NASSDD381 pKa = 3.23VGEE384 pKa = 4.37FVEE387 pKa = 4.41IAGPAGTDD395 pKa = 3.27LTGVSIEE402 pKa = 4.64LYY404 pKa = 9.51NGNGGGVYY412 pKa = 8.68KK413 pKa = 10.27TVALSGTIPEE423 pKa = 4.17QAGGFGTVAEE433 pKa = 4.82YY434 pKa = 10.41ISGIQNGAPDD444 pKa = 4.66GVALSYY450 pKa = 10.93NGEE453 pKa = 4.39LIDD456 pKa = 4.44FISYY460 pKa = 10.18EE461 pKa = 4.32GEE463 pKa = 3.82LTATGGSASGVTATDD478 pKa = 2.63IGVAEE483 pKa = 4.38TGSTLSTEE491 pKa = 4.58SVQLTGTGSEE501 pKa = 4.21VCHH504 pKa = 6.31FSWSGPATSSPGEE517 pKa = 3.97INAGQTFDD525 pKa = 3.67ASVGATCGGNDD536 pKa = 4.05GGDD539 pKa = 3.29GGTGGEE545 pKa = 4.19VTLVKK550 pKa = 10.18ISEE553 pKa = 4.38VQGSGSSSPLINAQVAIEE571 pKa = 4.64AIVVADD577 pKa = 3.97HH578 pKa = 6.24QSGSGTDD585 pKa = 3.18GDD587 pKa = 3.86LRR589 pKa = 11.84GFFVQEE595 pKa = 3.56EE596 pKa = 4.23DD597 pKa = 3.65ADD599 pKa = 4.06ADD601 pKa = 4.07ADD603 pKa = 3.95SSTSEE608 pKa = 3.9GLFVFDD614 pKa = 4.65GSSPAVDD621 pKa = 3.33VAVGDD626 pKa = 3.67KK627 pKa = 10.6VYY629 pKa = 11.07LVGTVDD635 pKa = 3.99EE636 pKa = 4.82YY637 pKa = 11.51FGDD640 pKa = 3.78TQLDD644 pKa = 3.95VQSGSVQVVSSGNALPSPAIVTLPLATTTNSGGEE678 pKa = 4.12AIADD682 pKa = 3.98LEE684 pKa = 4.61AYY686 pKa = 10.0EE687 pKa = 4.61GMQVMFAQTLAVTEE701 pKa = 4.48LFNLDD706 pKa = 3.23RR707 pKa = 11.84FGEE710 pKa = 4.15ILLSSEE716 pKa = 3.9GRR718 pKa = 11.84QVQYY722 pKa = 10.61TEE724 pKa = 3.61IMTPSVEE731 pKa = 4.28GYY733 pKa = 9.5AAYY736 pKa = 10.23VASAAANRR744 pKa = 11.84LMLDD748 pKa = 3.95DD749 pKa = 5.41GFSIQNPSPIRR760 pKa = 11.84FPAPGLSTANTVRR773 pKa = 11.84MSDD776 pKa = 3.18EE777 pKa = 4.18VTNLTGVISYY787 pKa = 10.3RR788 pKa = 11.84RR789 pKa = 11.84GSGGSGDD796 pKa = 3.3EE797 pKa = 4.64MYY799 pKa = 11.26RR800 pKa = 11.84LMPTVEE806 pKa = 4.2PVFTRR811 pKa = 11.84PEE813 pKa = 4.0SRR815 pKa = 11.84KK816 pKa = 9.97DD817 pKa = 3.5RR818 pKa = 11.84PDD820 pKa = 3.13VGGSIQVASFNLLNFFNTFGNSCYY844 pKa = 10.41LGGGTGDD851 pKa = 4.61CRR853 pKa = 11.84GADD856 pKa = 3.3NTEE859 pKa = 3.96EE860 pKa = 4.72YY861 pKa = 10.73DD862 pKa = 3.59RR863 pKa = 11.84QLQKK867 pKa = 9.98TVTAITALDD876 pKa = 3.83ADD878 pKa = 3.99VLGVIEE884 pKa = 4.5IEE886 pKa = 3.86NDD888 pKa = 2.9IVEE891 pKa = 4.61GQNSAMASLVNALNQAGSACSDD913 pKa = 3.55YY914 pKa = 11.19QYY916 pKa = 11.28VDD918 pKa = 3.16PQQRR922 pKa = 11.84VGTDD926 pKa = 3.95AIAVGFLYY934 pKa = 10.84CADD937 pKa = 3.9TVEE940 pKa = 4.46VTAGSSIAILDD951 pKa = 3.99DD952 pKa = 3.73SQLAGLGLSDD962 pKa = 4.09YY963 pKa = 11.42APVFDD968 pKa = 5.04GSSTSRR974 pKa = 11.84PSLTVSFTEE983 pKa = 4.17KK984 pKa = 9.81ATSEE988 pKa = 4.27VFTVSVNHH996 pKa = 6.4FKK998 pKa = 11.19SKK1000 pKa = 10.51GGSGSGGNADD1010 pKa = 4.34ANDD1013 pKa = 4.2GAGNWNEE1020 pKa = 3.8RR1021 pKa = 11.84RR1022 pKa = 11.84TQAAEE1027 pKa = 4.13ALHH1030 pKa = 6.59KK1031 pKa = 10.11WLATYY1036 pKa = 7.59PTGVSDD1042 pKa = 6.11DD1043 pKa = 5.08DD1044 pKa = 3.59ILIIGDD1050 pKa = 4.08LNAYY1054 pKa = 10.38SMEE1057 pKa = 4.26DD1058 pKa = 3.69PIVEE1062 pKa = 4.23LTSNGYY1068 pKa = 8.01TNVTGGEE1075 pKa = 3.67AHH1077 pKa = 6.19YY1078 pKa = 10.81SYY1080 pKa = 11.49VFDD1083 pKa = 4.39GFAGNLDD1090 pKa = 3.84HH1091 pKa = 7.1GLASASLMDD1100 pKa = 4.6QITGATSWNINSDD1113 pKa = 3.36EE1114 pKa = 4.75ADD1116 pKa = 3.46ALDD1119 pKa = 3.78YY1120 pKa = 10.35NTDD1123 pKa = 3.76FGRR1126 pKa = 11.84PTDD1129 pKa = 3.76IFDD1132 pKa = 3.45GTTPYY1137 pKa = 9.19RR1138 pKa = 11.84TSDD1141 pKa = 3.79HH1142 pKa = 7.29DD1143 pKa = 3.93PLLIGLSLSSVLKK1156 pKa = 10.79GDD1158 pKa = 3.97LDD1160 pKa = 4.21GDD1162 pKa = 4.04GDD1164 pKa = 4.11IDD1166 pKa = 4.79RR1167 pKa = 11.84MDD1169 pKa = 5.65FILIMRR1175 pKa = 11.84AILTRR1180 pKa = 11.84KK1181 pKa = 9.6PFDD1184 pKa = 3.48AANDD1188 pKa = 3.65INGDD1192 pKa = 3.78GVVNIVDD1199 pKa = 3.65GLLLRR1204 pKa = 11.84RR1205 pKa = 11.84SCTRR1209 pKa = 11.84ARR1211 pKa = 11.84CAVRR1215 pKa = 4.07
Molecular weight: 125.84 kDa
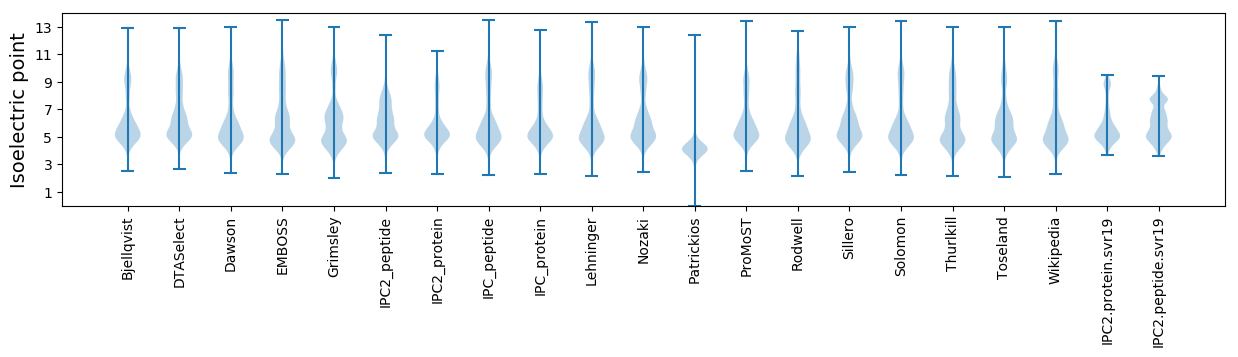
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1M5GDS0|A0A1M5GDS0_9ALTE Endonuclease III OS=Marisediminitalea aggregata OX=634436 GN=nth PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.14RR12 pKa = 11.84KK13 pKa = 9.13RR14 pKa = 11.84SHH16 pKa = 6.17GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37NGRR28 pKa = 11.84KK29 pKa = 9.16VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.29GRR39 pKa = 11.84ARR41 pKa = 11.84LSAA44 pKa = 3.91
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.14RR12 pKa = 11.84KK13 pKa = 9.13RR14 pKa = 11.84SHH16 pKa = 6.17GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37NGRR28 pKa = 11.84KK29 pKa = 9.16VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.29GRR39 pKa = 11.84ARR41 pKa = 11.84LSAA44 pKa = 3.91
Molecular weight: 5.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1469709 |
29 |
2683 |
340.8 |
37.69 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.415 ± 0.039 | 0.99 ± 0.012 |
5.801 ± 0.031 | 5.902 ± 0.035 |
4.138 ± 0.025 | 6.895 ± 0.038 |
2.229 ± 0.021 | 5.789 ± 0.027 |
4.485 ± 0.03 | 10.138 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.493 ± 0.016 | 4.185 ± 0.027 |
4.159 ± 0.022 | 4.717 ± 0.032 |
4.625 ± 0.026 | 6.577 ± 0.029 |
5.729 ± 0.03 | 7.229 ± 0.032 |
1.327 ± 0.013 | 3.179 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
