
Flavimobilis marinus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micro
Average proteome isoelectric point is 6.02
Get precalculated fractions of proteins
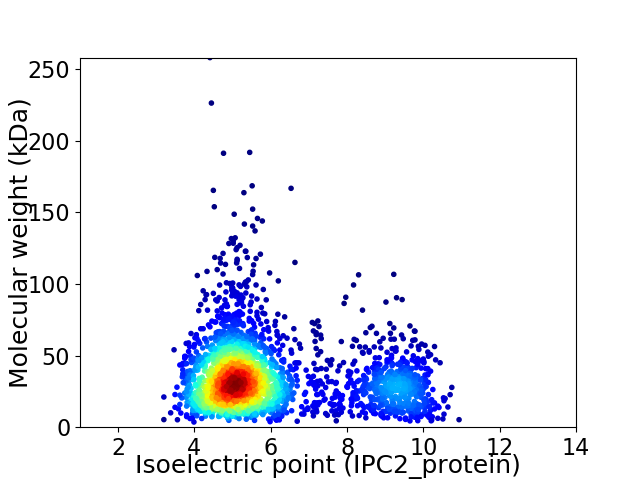
Virtual 2D-PAGE plot for 2643 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I2GLJ4|A0A1I2GLJ4_9MICO Chemotaxis phosphatase CheX OS=Flavimobilis marinus OX=285351 GN=SAMN04488035_1851 PE=4 SV=1
MM1 pKa = 6.96TAAGTPRR8 pKa = 11.84RR9 pKa = 11.84SLRR12 pKa = 11.84AVGAIAALGLVLTACADD29 pKa = 3.56TSEE32 pKa = 5.38GDD34 pKa = 3.86DD35 pKa = 3.56PTATGGTAGGGTDD48 pKa = 4.0SISVGTTDD56 pKa = 4.96VITSLDD62 pKa = 3.58PAGSYY67 pKa = 11.34DD68 pKa = 3.46NGSFAVQNQVFPFLLNTPYY87 pKa = 10.83GSPDD91 pKa = 3.56VEE93 pKa = 3.89PDD95 pKa = 3.01IAEE98 pKa = 4.28SAEE101 pKa = 3.81FTAPNEE107 pKa = 3.95YY108 pKa = 9.34TVTLKK113 pKa = 10.98EE114 pKa = 4.14GLTFANGHH122 pKa = 6.04EE123 pKa = 4.48LTSSDD128 pKa = 3.4VKK130 pKa = 10.92FSFDD134 pKa = 3.51RR135 pKa = 11.84QLGIADD141 pKa = 3.9PNGPSSLLYY150 pKa = 10.92NLDD153 pKa = 3.31SVAAPDD159 pKa = 3.39EE160 pKa = 4.27RR161 pKa = 11.84TVVFTLKK168 pKa = 10.58SEE170 pKa = 4.16NDD172 pKa = 3.17QTFPQILSSPVGPIVDD188 pKa = 3.95EE189 pKa = 4.62EE190 pKa = 4.69VFSPTEE196 pKa = 3.84LTPADD201 pKa = 3.76TIVAEE206 pKa = 4.39KK207 pKa = 10.95AFGGQYY213 pKa = 10.75LITDD217 pKa = 3.57YY218 pKa = 11.33TEE220 pKa = 4.27NEE222 pKa = 4.42LIRR225 pKa = 11.84YY226 pKa = 7.48EE227 pKa = 4.13AFEE230 pKa = 5.97DD231 pKa = 4.04YY232 pKa = 10.32MGLLEE237 pKa = 4.19PAVTKK242 pKa = 10.71VVTAQYY248 pKa = 8.37FTEE251 pKa = 4.25EE252 pKa = 4.15TSLKK256 pKa = 10.64LAVQQGDD263 pKa = 3.15VDD265 pKa = 3.86VAFRR269 pKa = 11.84SLSPTDD275 pKa = 4.08LADD278 pKa = 4.08LAQDD282 pKa = 3.57EE283 pKa = 4.74NVTVHH288 pKa = 6.82EE289 pKa = 5.04GPGGEE294 pKa = 3.43IRR296 pKa = 11.84YY297 pKa = 9.32IVFNFATQPYY307 pKa = 8.21GTDD310 pKa = 2.91TADD313 pKa = 3.45ADD315 pKa = 3.92EE316 pKa = 4.82AKK318 pKa = 10.76ALAVRR323 pKa = 11.84QAAAHH328 pKa = 5.97LLDD331 pKa = 3.81RR332 pKa = 11.84QALSEE337 pKa = 4.41EE338 pKa = 4.78IYY340 pKa = 10.59QGSFTPLYY348 pKa = 10.57SYY350 pKa = 11.35VPDD353 pKa = 4.28GLTGAAEE360 pKa = 4.12PLKK363 pKa = 10.47EE364 pKa = 4.39LYY366 pKa = 10.64GDD368 pKa = 3.79GAGAPDD374 pKa = 4.2ADD376 pKa = 3.81AAAAALSEE384 pKa = 4.67AGVEE388 pKa = 4.35TPVALNLQYY397 pKa = 11.28SPDD400 pKa = 3.57HH401 pKa = 6.48YY402 pKa = 11.26GNSSDD407 pKa = 4.48EE408 pKa = 4.18EE409 pKa = 4.34YY410 pKa = 11.85AMIEE414 pKa = 4.13AQLEE418 pKa = 4.08ASGLFDD424 pKa = 4.59VSLQSTLWDD433 pKa = 3.61TYY435 pKa = 7.98STEE438 pKa = 3.86RR439 pKa = 11.84RR440 pKa = 11.84EE441 pKa = 4.39DD442 pKa = 3.43VYY444 pKa = 10.88PAYY447 pKa = 10.28QLGWFPDD454 pKa = 3.67YY455 pKa = 11.48SDD457 pKa = 3.97ADD459 pKa = 3.75NYY461 pKa = 11.0LSPFFLPEE469 pKa = 3.76NFLGNHH475 pKa = 5.81YY476 pKa = 10.32EE477 pKa = 4.43DD478 pKa = 5.09DD479 pKa = 4.03AATEE483 pKa = 4.83AILGQASEE491 pKa = 4.35ADD493 pKa = 3.57EE494 pKa = 4.39AARR497 pKa = 11.84TALIEE502 pKa = 4.18DD503 pKa = 3.77AQRR506 pKa = 11.84IVAEE510 pKa = 4.58DD511 pKa = 4.54LSTLPYY517 pKa = 10.46LQGAQVAVAGTDD529 pKa = 3.21ITGVTLDD536 pKa = 3.42ASFKK540 pKa = 10.46FRR542 pKa = 11.84YY543 pKa = 9.76APLGRR548 pKa = 4.77
MM1 pKa = 6.96TAAGTPRR8 pKa = 11.84RR9 pKa = 11.84SLRR12 pKa = 11.84AVGAIAALGLVLTACADD29 pKa = 3.56TSEE32 pKa = 5.38GDD34 pKa = 3.86DD35 pKa = 3.56PTATGGTAGGGTDD48 pKa = 4.0SISVGTTDD56 pKa = 4.96VITSLDD62 pKa = 3.58PAGSYY67 pKa = 11.34DD68 pKa = 3.46NGSFAVQNQVFPFLLNTPYY87 pKa = 10.83GSPDD91 pKa = 3.56VEE93 pKa = 3.89PDD95 pKa = 3.01IAEE98 pKa = 4.28SAEE101 pKa = 3.81FTAPNEE107 pKa = 3.95YY108 pKa = 9.34TVTLKK113 pKa = 10.98EE114 pKa = 4.14GLTFANGHH122 pKa = 6.04EE123 pKa = 4.48LTSSDD128 pKa = 3.4VKK130 pKa = 10.92FSFDD134 pKa = 3.51RR135 pKa = 11.84QLGIADD141 pKa = 3.9PNGPSSLLYY150 pKa = 10.92NLDD153 pKa = 3.31SVAAPDD159 pKa = 3.39EE160 pKa = 4.27RR161 pKa = 11.84TVVFTLKK168 pKa = 10.58SEE170 pKa = 4.16NDD172 pKa = 3.17QTFPQILSSPVGPIVDD188 pKa = 3.95EE189 pKa = 4.62EE190 pKa = 4.69VFSPTEE196 pKa = 3.84LTPADD201 pKa = 3.76TIVAEE206 pKa = 4.39KK207 pKa = 10.95AFGGQYY213 pKa = 10.75LITDD217 pKa = 3.57YY218 pKa = 11.33TEE220 pKa = 4.27NEE222 pKa = 4.42LIRR225 pKa = 11.84YY226 pKa = 7.48EE227 pKa = 4.13AFEE230 pKa = 5.97DD231 pKa = 4.04YY232 pKa = 10.32MGLLEE237 pKa = 4.19PAVTKK242 pKa = 10.71VVTAQYY248 pKa = 8.37FTEE251 pKa = 4.25EE252 pKa = 4.15TSLKK256 pKa = 10.64LAVQQGDD263 pKa = 3.15VDD265 pKa = 3.86VAFRR269 pKa = 11.84SLSPTDD275 pKa = 4.08LADD278 pKa = 4.08LAQDD282 pKa = 3.57EE283 pKa = 4.74NVTVHH288 pKa = 6.82EE289 pKa = 5.04GPGGEE294 pKa = 3.43IRR296 pKa = 11.84YY297 pKa = 9.32IVFNFATQPYY307 pKa = 8.21GTDD310 pKa = 2.91TADD313 pKa = 3.45ADD315 pKa = 3.92EE316 pKa = 4.82AKK318 pKa = 10.76ALAVRR323 pKa = 11.84QAAAHH328 pKa = 5.97LLDD331 pKa = 3.81RR332 pKa = 11.84QALSEE337 pKa = 4.41EE338 pKa = 4.78IYY340 pKa = 10.59QGSFTPLYY348 pKa = 10.57SYY350 pKa = 11.35VPDD353 pKa = 4.28GLTGAAEE360 pKa = 4.12PLKK363 pKa = 10.47EE364 pKa = 4.39LYY366 pKa = 10.64GDD368 pKa = 3.79GAGAPDD374 pKa = 4.2ADD376 pKa = 3.81AAAAALSEE384 pKa = 4.67AGVEE388 pKa = 4.35TPVALNLQYY397 pKa = 11.28SPDD400 pKa = 3.57HH401 pKa = 6.48YY402 pKa = 11.26GNSSDD407 pKa = 4.48EE408 pKa = 4.18EE409 pKa = 4.34YY410 pKa = 11.85AMIEE414 pKa = 4.13AQLEE418 pKa = 4.08ASGLFDD424 pKa = 4.59VSLQSTLWDD433 pKa = 3.61TYY435 pKa = 7.98STEE438 pKa = 3.86RR439 pKa = 11.84RR440 pKa = 11.84EE441 pKa = 4.39DD442 pKa = 3.43VYY444 pKa = 10.88PAYY447 pKa = 10.28QLGWFPDD454 pKa = 3.67YY455 pKa = 11.48SDD457 pKa = 3.97ADD459 pKa = 3.75NYY461 pKa = 11.0LSPFFLPEE469 pKa = 3.76NFLGNHH475 pKa = 5.81YY476 pKa = 10.32EE477 pKa = 4.43DD478 pKa = 5.09DD479 pKa = 4.03AATEE483 pKa = 4.83AILGQASEE491 pKa = 4.35ADD493 pKa = 3.57EE494 pKa = 4.39AARR497 pKa = 11.84TALIEE502 pKa = 4.18DD503 pKa = 3.77AQRR506 pKa = 11.84IVAEE510 pKa = 4.58DD511 pKa = 4.54LSTLPYY517 pKa = 10.46LQGAQVAVAGTDD529 pKa = 3.21ITGVTLDD536 pKa = 3.42ASFKK540 pKa = 10.46FRR542 pKa = 11.84YY543 pKa = 9.76APLGRR548 pKa = 4.77
Molecular weight: 58.51 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I2HW18|A0A1I2HW18_9MICO Ribosomal protein S18 acetylase RimI OS=Flavimobilis marinus OX=285351 GN=SAMN04488035_2546 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.16GFRR20 pKa = 11.84LRR22 pKa = 11.84MSTRR26 pKa = 11.84AGRR29 pKa = 11.84AILAARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.65GRR40 pKa = 11.84SEE42 pKa = 3.94LSAA45 pKa = 4.73
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.16GFRR20 pKa = 11.84LRR22 pKa = 11.84MSTRR26 pKa = 11.84AGRR29 pKa = 11.84AILAARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.65GRR40 pKa = 11.84SEE42 pKa = 3.94LSAA45 pKa = 4.73
Molecular weight: 5.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
894391 |
33 |
2433 |
338.4 |
35.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.351 ± 0.074 | 0.56 ± 0.011 |
6.319 ± 0.039 | 5.647 ± 0.045 |
2.63 ± 0.03 | 9.062 ± 0.043 |
2.07 ± 0.024 | 3.417 ± 0.034 |
1.8 ± 0.036 | 10.208 ± 0.059 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.749 ± 0.019 | 1.622 ± 0.026 |
5.628 ± 0.038 | 2.714 ± 0.023 |
7.589 ± 0.048 | 5.144 ± 0.031 |
6.324 ± 0.044 | 9.855 ± 0.054 |
1.445 ± 0.018 | 1.868 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
