
Botryosphaeria parva (strain UCR-NP2) (Grapevine canker fungus) (Neofusicoccum parvum)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; dothideomyceta; Dothideomycetes; Dothideomycetes incertae sedis; Botryosphaeriales; Botryosphaeriaceae; Neofusicoccum; Neofusicoccum parvum
Average proteome isoelectric point is 6.25
Get precalculated fractions of proteins
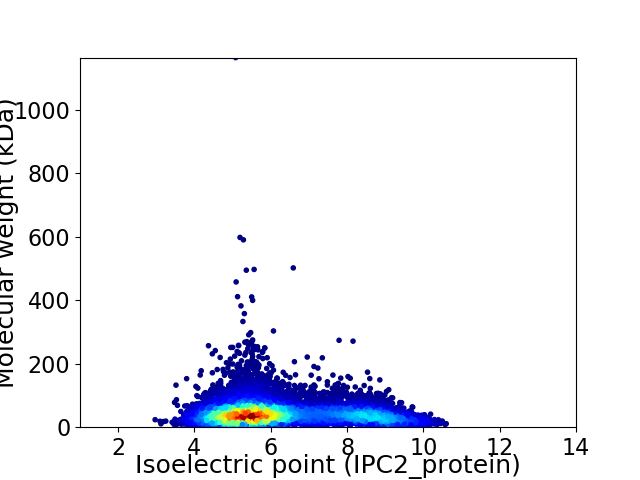
Virtual 2D-PAGE plot for 10365 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
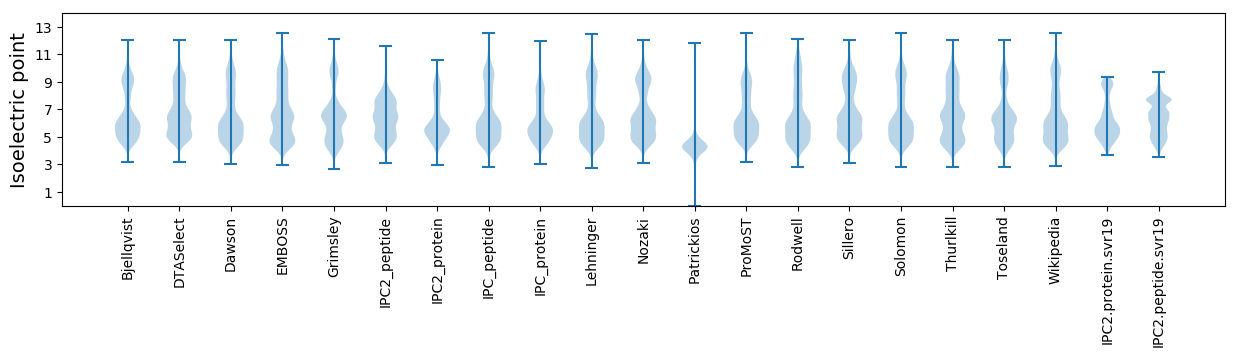
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R1ERX5|R1ERX5_BOTPV Putative outer membrane autotransporter protein OS=Botryosphaeria parva (strain UCR-NP2) OX=1287680 GN=UCRNP2_2714 PE=4 SV=1
MM1 pKa = 7.46FFNILFDD8 pKa = 3.84LFFNVFFDD16 pKa = 3.6IFLDD20 pKa = 3.35MFFNIFFEE28 pKa = 4.12IFFDD32 pKa = 3.14ILLNIFFNFFPNILLSIFLNVFDD55 pKa = 4.46IFFNIFSYY63 pKa = 10.74IVFDD67 pKa = 4.03TVFGASTTISDD78 pKa = 3.95LLIPVFAFFTVIYY91 pKa = 9.97FFDD94 pKa = 3.57ILPRR98 pKa = 11.84LLLHH102 pKa = 7.08AFNIPDD108 pKa = 4.01TLLSTLDD115 pKa = 3.69FFDD118 pKa = 3.37ILHH121 pKa = 6.68RR122 pKa = 11.84LFSIFQLSQATIFLNLYY139 pKa = 9.88FSRR142 pKa = 11.84LCFII146 pKa = 5.38
MM1 pKa = 7.46FFNILFDD8 pKa = 3.84LFFNVFFDD16 pKa = 3.6IFLDD20 pKa = 3.35MFFNIFFEE28 pKa = 4.12IFFDD32 pKa = 3.14ILLNIFFNFFPNILLSIFLNVFDD55 pKa = 4.46IFFNIFSYY63 pKa = 10.74IVFDD67 pKa = 4.03TVFGASTTISDD78 pKa = 3.95LLIPVFAFFTVIYY91 pKa = 9.97FFDD94 pKa = 3.57ILPRR98 pKa = 11.84LLLHH102 pKa = 7.08AFNIPDD108 pKa = 4.01TLLSTLDD115 pKa = 3.69FFDD118 pKa = 3.37ILHH121 pKa = 6.68RR122 pKa = 11.84LFSIFQLSQATIFLNLYY139 pKa = 9.88FSRR142 pKa = 11.84LCFII146 pKa = 5.38
Molecular weight: 17.6 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R1EK03|R1EK03_BOTPV Putative transcription initiation factor 31kd protein OS=Botryosphaeria parva (strain UCR-NP2) OX=1287680 GN=UCRNP2_5394 PE=3 SV=1
MM1 pKa = 7.6RR2 pKa = 11.84PSPRR6 pKa = 11.84LWAFNAYY13 pKa = 9.88CSVHH17 pKa = 4.85TVLYY21 pKa = 10.41CRR23 pKa = 11.84ARR25 pKa = 11.84NSAPAFGVGLVSAWSLLWTFTLLVVHH51 pKa = 7.34DD52 pKa = 4.73AQTDD56 pKa = 4.04FARR59 pKa = 11.84IEE61 pKa = 4.16RR62 pKa = 11.84TEE64 pKa = 4.04GASASLRR71 pKa = 11.84ISNGSATPNGSAQNGKK87 pKa = 8.52QEE89 pKa = 4.07QEE91 pKa = 3.85SRR93 pKa = 11.84AVSTIKK99 pKa = 10.91DD100 pKa = 3.04VDD102 pKa = 3.82LAAEE106 pKa = 4.23RR107 pKa = 11.84KK108 pKa = 9.91KK109 pKa = 11.04LGTTAGPTQRR119 pKa = 11.84RR120 pKa = 11.84GTFAWQHH127 pKa = 5.44YY128 pKa = 8.12PLSPFIEE135 pKa = 4.33RR136 pKa = 11.84LDD138 pKa = 3.45WVADD142 pKa = 3.42VFCNFRR148 pKa = 11.84GMGWNWRR155 pKa = 11.84ISGIAPPPRR164 pKa = 11.84WVQQQLHH171 pKa = 6.47EE172 pKa = 4.35NSGTPISSTPDD183 pKa = 2.88NHH185 pKa = 6.97VGVDD189 pKa = 3.68GTVQPQTRR197 pKa = 11.84SEE199 pKa = 3.97ALRR202 pKa = 11.84AGWRR206 pKa = 11.84TFITGYY212 pKa = 10.35LALDD216 pKa = 3.88LLKK219 pKa = 10.83VITIHH224 pKa = 7.61DD225 pKa = 4.2PYY227 pKa = 11.15FWGLTTAPGPSFLPPFIRR245 pKa = 11.84HH246 pKa = 5.78SPTLLRR252 pKa = 11.84TFRR255 pKa = 11.84LIVSLFAIKK264 pKa = 8.98TALTTIFAMAPLFFIGLLGPRR285 pKa = 11.84AIGARR290 pKa = 11.84ASPWMYY296 pKa = 10.19PDD298 pKa = 3.29TFGSFRR304 pKa = 11.84SVLDD308 pKa = 3.51HH309 pKa = 7.16GLAGWWGSWWHH320 pKa = 4.2QTFRR324 pKa = 11.84FAFSAPSALLARR336 pKa = 11.84SLGLDD341 pKa = 3.01RR342 pKa = 11.84RR343 pKa = 11.84GAPARR348 pKa = 11.84ALQLLVAFALSGCLHH363 pKa = 6.88AAGSHH368 pKa = 4.5TQAGATRR375 pKa = 11.84PLSSFLFFVLQAAAIVAQQALARR398 pKa = 11.84VPVVRR403 pKa = 11.84RR404 pKa = 11.84VPKK407 pKa = 10.43GVRR410 pKa = 11.84QLANFVVVHH419 pKa = 4.92VWFYY423 pKa = 9.56YY424 pKa = 8.2TAPLLADD431 pKa = 4.41DD432 pKa = 5.11FAHH435 pKa = 6.49GGIWLFEE442 pKa = 4.15PVPLSPLRR450 pKa = 11.84GMLGLGGKK458 pKa = 8.39HH459 pKa = 4.85WWEE462 pKa = 3.85SGIAFF467 pKa = 4.8
MM1 pKa = 7.6RR2 pKa = 11.84PSPRR6 pKa = 11.84LWAFNAYY13 pKa = 9.88CSVHH17 pKa = 4.85TVLYY21 pKa = 10.41CRR23 pKa = 11.84ARR25 pKa = 11.84NSAPAFGVGLVSAWSLLWTFTLLVVHH51 pKa = 7.34DD52 pKa = 4.73AQTDD56 pKa = 4.04FARR59 pKa = 11.84IEE61 pKa = 4.16RR62 pKa = 11.84TEE64 pKa = 4.04GASASLRR71 pKa = 11.84ISNGSATPNGSAQNGKK87 pKa = 8.52QEE89 pKa = 4.07QEE91 pKa = 3.85SRR93 pKa = 11.84AVSTIKK99 pKa = 10.91DD100 pKa = 3.04VDD102 pKa = 3.82LAAEE106 pKa = 4.23RR107 pKa = 11.84KK108 pKa = 9.91KK109 pKa = 11.04LGTTAGPTQRR119 pKa = 11.84RR120 pKa = 11.84GTFAWQHH127 pKa = 5.44YY128 pKa = 8.12PLSPFIEE135 pKa = 4.33RR136 pKa = 11.84LDD138 pKa = 3.45WVADD142 pKa = 3.42VFCNFRR148 pKa = 11.84GMGWNWRR155 pKa = 11.84ISGIAPPPRR164 pKa = 11.84WVQQQLHH171 pKa = 6.47EE172 pKa = 4.35NSGTPISSTPDD183 pKa = 2.88NHH185 pKa = 6.97VGVDD189 pKa = 3.68GTVQPQTRR197 pKa = 11.84SEE199 pKa = 3.97ALRR202 pKa = 11.84AGWRR206 pKa = 11.84TFITGYY212 pKa = 10.35LALDD216 pKa = 3.88LLKK219 pKa = 10.83VITIHH224 pKa = 7.61DD225 pKa = 4.2PYY227 pKa = 11.15FWGLTTAPGPSFLPPFIRR245 pKa = 11.84HH246 pKa = 5.78SPTLLRR252 pKa = 11.84TFRR255 pKa = 11.84LIVSLFAIKK264 pKa = 8.98TALTTIFAMAPLFFIGLLGPRR285 pKa = 11.84AIGARR290 pKa = 11.84ASPWMYY296 pKa = 10.19PDD298 pKa = 3.29TFGSFRR304 pKa = 11.84SVLDD308 pKa = 3.51HH309 pKa = 7.16GLAGWWGSWWHH320 pKa = 4.2QTFRR324 pKa = 11.84FAFSAPSALLARR336 pKa = 11.84SLGLDD341 pKa = 3.01RR342 pKa = 11.84RR343 pKa = 11.84GAPARR348 pKa = 11.84ALQLLVAFALSGCLHH363 pKa = 6.88AAGSHH368 pKa = 4.5TQAGATRR375 pKa = 11.84PLSSFLFFVLQAAAIVAQQALARR398 pKa = 11.84VPVVRR403 pKa = 11.84RR404 pKa = 11.84VPKK407 pKa = 10.43GVRR410 pKa = 11.84QLANFVVVHH419 pKa = 4.92VWFYY423 pKa = 9.56YY424 pKa = 8.2TAPLLADD431 pKa = 4.41DD432 pKa = 5.11FAHH435 pKa = 6.49GGIWLFEE442 pKa = 4.15PVPLSPLRR450 pKa = 11.84GMLGLGGKK458 pKa = 8.39HH459 pKa = 4.85WWEE462 pKa = 3.85SGIAFF467 pKa = 4.8
Molecular weight: 51.5 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4404707 |
66 |
10721 |
425.0 |
46.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.98 ± 0.03 | 1.15 ± 0.01 |
5.903 ± 0.021 | 6.249 ± 0.028 |
3.841 ± 0.016 | 7.324 ± 0.023 |
2.272 ± 0.01 | 4.604 ± 0.017 |
4.852 ± 0.025 | 8.75 ± 0.022 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.167 ± 0.01 | 3.542 ± 0.016 |
5.759 ± 0.022 | 3.77 ± 0.015 |
5.822 ± 0.022 | 7.284 ± 0.024 |
5.854 ± 0.017 | 6.474 ± 0.021 |
1.537 ± 0.009 | 2.867 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
