
Macacine gammaherpesvirus 4 (Rhesus lymphocryptovirus)
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Peploviricota; Herviviricetes; Herpesvirales; Herpesviridae; Gammaherpesvirinae; Lymphocryptovirus
Average proteome isoelectric point is 6.91
Get precalculated fractions of proteins
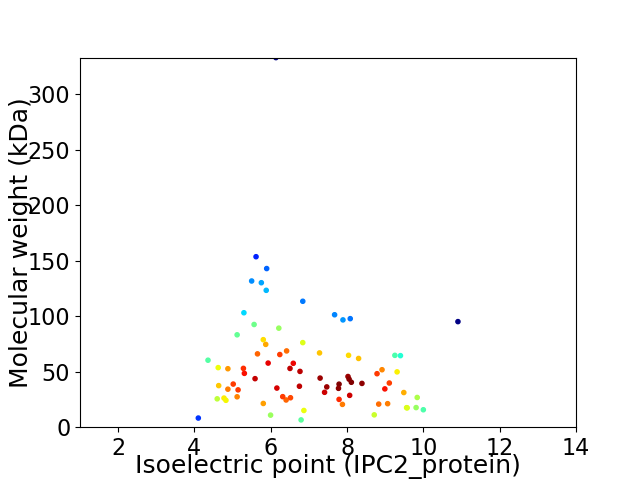
Virtual 2D-PAGE plot for 80 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q8UZC8|Q8UZC8_9GAMA BNLF2a OS=Macacine gammaherpesvirus 4 OX=45455 PE=4 SV=1
MM1 pKa = 7.49EE2 pKa = 5.08GNRR5 pKa = 11.84GRR7 pKa = 11.84GGGHH11 pKa = 5.58RR12 pKa = 11.84HH13 pKa = 5.63PRR15 pKa = 11.84CPQPHH20 pKa = 6.79ASLLFGSLLLLVAILVWFFIIMSDD44 pKa = 3.82LTQTASTVLSSFAVVLIIIIIIIMLFKK71 pKa = 10.67RR72 pKa = 11.84RR73 pKa = 11.84LLCPLGLLCIILIMIVILVSNLLTLTGQTLFVGLVILCLCILLALAVWFYY123 pKa = 11.4LMWLLRR129 pKa = 11.84EE130 pKa = 4.12YY131 pKa = 10.72GASFWTILAFCLAFLLAIVFLIIAVLLHH159 pKa = 6.18LAWFTVLVDD168 pKa = 5.16LYY170 pKa = 10.68WLLLFLAILIWIYY183 pKa = 11.02VHH185 pKa = 7.04DD186 pKa = 4.89FNRR189 pKa = 11.84AIPEE193 pKa = 3.88ALANLEE199 pKa = 4.13EE200 pKa = 4.73EE201 pKa = 4.88YY202 pKa = 11.04NHH204 pKa = 6.37QGHH207 pKa = 5.88GHH209 pKa = 6.58GGGQGDD215 pKa = 3.61EE216 pKa = 4.19HH217 pKa = 7.07QPRR220 pKa = 11.84VLLISEE226 pKa = 4.35DD227 pKa = 3.85PKK229 pKa = 9.55THH231 pKa = 6.08HH232 pKa = 6.64HH233 pKa = 6.22HH234 pKa = 6.49HH235 pKa = 6.59HH236 pKa = 6.16HH237 pKa = 6.2HH238 pKa = 5.78HH239 pKa = 6.83HH240 pKa = 6.51KK241 pKa = 10.64NSNQGDD247 pKa = 3.81PLGPYY252 pKa = 9.87VSQNGGNGGNEE263 pKa = 3.69GDD265 pKa = 3.78GGNRR269 pKa = 11.84DD270 pKa = 3.63GGNGDD275 pKa = 3.98GGNGDD280 pKa = 4.1GGNVGDD286 pKa = 4.76GGNGNGGNGDD296 pKa = 3.9GGNGDD301 pKa = 4.23GGNGDD306 pKa = 4.62GGNEE310 pKa = 4.43GPDD313 pKa = 3.37HH314 pKa = 6.93LPYY317 pKa = 10.35PIQATDD323 pKa = 3.26GGNGGEE329 pKa = 3.95GDD331 pKa = 4.35GGNEE335 pKa = 4.32GPDD338 pKa = 3.61PLPYY342 pKa = 10.11PIQATDD348 pKa = 3.62GGNGGQGDD356 pKa = 4.21GGDD359 pKa = 3.7GDD361 pKa = 4.54GGNGGNGGGGGDD373 pKa = 4.57PDD375 pKa = 4.75HH376 pKa = 7.06LPHH379 pKa = 7.6PIQATDD385 pKa = 3.53GGNSGDD391 pKa = 4.38GGDD394 pKa = 4.51GGDD397 pKa = 4.2GEE399 pKa = 5.03DD400 pKa = 4.6GEE402 pKa = 5.17GGGDD406 pKa = 3.22RR407 pKa = 11.84GPFGPYY413 pKa = 9.56VSQGPSGDD421 pKa = 4.24PDD423 pKa = 4.0HH424 pKa = 7.07LPHH427 pKa = 7.16PVQASDD433 pKa = 3.76GGDD436 pKa = 3.14GLGPYY441 pKa = 9.93GPAGPFTQGPSWPWGPFGTGPLGPWGPFGPGPCGPWGPWGPNNNHH486 pKa = 6.42GPLQEE491 pKa = 4.41TGPGGPWMLLTLGGGGNSVHH511 pKa = 6.94LNDD514 pKa = 5.28RR515 pKa = 11.84GNGGNGPQNPDD526 pKa = 2.93NSNGSGPGGPHH537 pKa = 5.25NQQPEE542 pKa = 4.44SNEE545 pKa = 3.96GGGPPGPTDD554 pKa = 3.78GGNNGPDD561 pKa = 3.71SPPSNQGPQGPEE573 pKa = 3.81GDD575 pKa = 4.47PGDD578 pKa = 4.28PADD581 pKa = 5.39PIQISYY587 pKa = 11.0YY588 pKa = 10.57DD589 pKa = 3.28
MM1 pKa = 7.49EE2 pKa = 5.08GNRR5 pKa = 11.84GRR7 pKa = 11.84GGGHH11 pKa = 5.58RR12 pKa = 11.84HH13 pKa = 5.63PRR15 pKa = 11.84CPQPHH20 pKa = 6.79ASLLFGSLLLLVAILVWFFIIMSDD44 pKa = 3.82LTQTASTVLSSFAVVLIIIIIIIMLFKK71 pKa = 10.67RR72 pKa = 11.84RR73 pKa = 11.84LLCPLGLLCIILIMIVILVSNLLTLTGQTLFVGLVILCLCILLALAVWFYY123 pKa = 11.4LMWLLRR129 pKa = 11.84EE130 pKa = 4.12YY131 pKa = 10.72GASFWTILAFCLAFLLAIVFLIIAVLLHH159 pKa = 6.18LAWFTVLVDD168 pKa = 5.16LYY170 pKa = 10.68WLLLFLAILIWIYY183 pKa = 11.02VHH185 pKa = 7.04DD186 pKa = 4.89FNRR189 pKa = 11.84AIPEE193 pKa = 3.88ALANLEE199 pKa = 4.13EE200 pKa = 4.73EE201 pKa = 4.88YY202 pKa = 11.04NHH204 pKa = 6.37QGHH207 pKa = 5.88GHH209 pKa = 6.58GGGQGDD215 pKa = 3.61EE216 pKa = 4.19HH217 pKa = 7.07QPRR220 pKa = 11.84VLLISEE226 pKa = 4.35DD227 pKa = 3.85PKK229 pKa = 9.55THH231 pKa = 6.08HH232 pKa = 6.64HH233 pKa = 6.22HH234 pKa = 6.49HH235 pKa = 6.59HH236 pKa = 6.16HH237 pKa = 6.2HH238 pKa = 5.78HH239 pKa = 6.83HH240 pKa = 6.51KK241 pKa = 10.64NSNQGDD247 pKa = 3.81PLGPYY252 pKa = 9.87VSQNGGNGGNEE263 pKa = 3.69GDD265 pKa = 3.78GGNRR269 pKa = 11.84DD270 pKa = 3.63GGNGDD275 pKa = 3.98GGNGDD280 pKa = 4.1GGNVGDD286 pKa = 4.76GGNGNGGNGDD296 pKa = 3.9GGNGDD301 pKa = 4.23GGNGDD306 pKa = 4.62GGNEE310 pKa = 4.43GPDD313 pKa = 3.37HH314 pKa = 6.93LPYY317 pKa = 10.35PIQATDD323 pKa = 3.26GGNGGEE329 pKa = 3.95GDD331 pKa = 4.35GGNEE335 pKa = 4.32GPDD338 pKa = 3.61PLPYY342 pKa = 10.11PIQATDD348 pKa = 3.62GGNGGQGDD356 pKa = 4.21GGDD359 pKa = 3.7GDD361 pKa = 4.54GGNGGNGGGGGDD373 pKa = 4.57PDD375 pKa = 4.75HH376 pKa = 7.06LPHH379 pKa = 7.6PIQATDD385 pKa = 3.53GGNSGDD391 pKa = 4.38GGDD394 pKa = 4.51GGDD397 pKa = 4.2GEE399 pKa = 5.03DD400 pKa = 4.6GEE402 pKa = 5.17GGGDD406 pKa = 3.22RR407 pKa = 11.84GPFGPYY413 pKa = 9.56VSQGPSGDD421 pKa = 4.24PDD423 pKa = 4.0HH424 pKa = 7.07LPHH427 pKa = 7.16PVQASDD433 pKa = 3.76GGDD436 pKa = 3.14GLGPYY441 pKa = 9.93GPAGPFTQGPSWPWGPFGTGPLGPWGPFGPGPCGPWGPWGPNNNHH486 pKa = 6.42GPLQEE491 pKa = 4.41TGPGGPWMLLTLGGGGNSVHH511 pKa = 6.94LNDD514 pKa = 5.28RR515 pKa = 11.84GNGGNGPQNPDD526 pKa = 2.93NSNGSGPGGPHH537 pKa = 5.25NQQPEE542 pKa = 4.44SNEE545 pKa = 3.96GGGPPGPTDD554 pKa = 3.78GGNNGPDD561 pKa = 3.71SPPSNQGPQGPEE573 pKa = 3.81GDD575 pKa = 4.47PGDD578 pKa = 4.28PADD581 pKa = 5.39PIQISYY587 pKa = 11.0YY588 pKa = 10.57DD589 pKa = 3.28
Molecular weight: 60.34 kDa
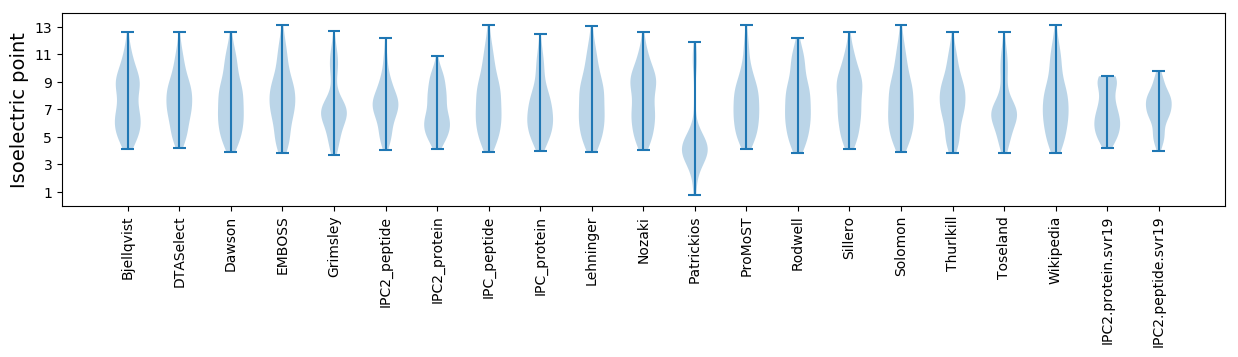
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q8UZE2|Q8UZE2_9GAMA BILF2 OS=Macacine gammaherpesvirus 4 OX=45455 PE=4 SV=1
MM1 pKa = 7.48GLGFRR6 pKa = 11.84GWGLHH11 pKa = 6.25EE12 pKa = 4.45PACLPGEE19 pKa = 4.31PNQRR23 pKa = 11.84TQGHH27 pKa = 6.82PAPGPSSRR35 pKa = 11.84TGQRR39 pKa = 11.84TRR41 pKa = 11.84QRR43 pKa = 11.84PGHH46 pKa = 6.35PPPEE50 pKa = 4.87RR51 pKa = 11.84GSGPRR56 pKa = 11.84GTRR59 pKa = 11.84PPAPSSRR66 pKa = 11.84TGQRR70 pKa = 11.84TRR72 pKa = 11.84QRR74 pKa = 11.84PGHH77 pKa = 6.35PPPEE81 pKa = 4.87RR82 pKa = 11.84GSGPRR87 pKa = 11.84GTRR90 pKa = 11.84PPAPSSRR97 pKa = 11.84TGQRR101 pKa = 11.84TRR103 pKa = 11.84QRR105 pKa = 11.84PGHH108 pKa = 6.35PPPEE112 pKa = 4.87RR113 pKa = 11.84GSGPRR118 pKa = 11.84GTRR121 pKa = 11.84PPAPSSRR128 pKa = 11.84TGQRR132 pKa = 11.84TRR134 pKa = 11.84QRR136 pKa = 11.84PGHH139 pKa = 6.35PPPEE143 pKa = 4.87RR144 pKa = 11.84GSGPRR149 pKa = 11.84GTRR152 pKa = 11.84PPAPSSRR159 pKa = 11.84TGQRR163 pKa = 11.84TRR165 pKa = 11.84QRR167 pKa = 11.84PGHH170 pKa = 6.35PPPEE174 pKa = 4.87RR175 pKa = 11.84GSGPRR180 pKa = 11.84GTRR183 pKa = 11.84PPAPSSRR190 pKa = 11.84TGQRR194 pKa = 11.84TRR196 pKa = 11.84QRR198 pKa = 11.84PGHH201 pKa = 6.35PPPEE205 pKa = 4.87RR206 pKa = 11.84GSGPRR211 pKa = 11.84GTRR214 pKa = 11.84PPAPSSRR221 pKa = 11.84TGQRR225 pKa = 11.84TRR227 pKa = 11.84QRR229 pKa = 11.84PGHH232 pKa = 6.35PPPEE236 pKa = 4.87RR237 pKa = 11.84GSGPRR242 pKa = 11.84GTRR245 pKa = 11.84PPAPSSRR252 pKa = 11.84TGQRR256 pKa = 11.84TRR258 pKa = 11.84QRR260 pKa = 11.84PGHH263 pKa = 6.35PPPEE267 pKa = 4.87RR268 pKa = 11.84GSGPRR273 pKa = 11.84GTRR276 pKa = 11.84PPAPSSRR283 pKa = 11.84TGQRR287 pKa = 11.84TRR289 pKa = 11.84QRR291 pKa = 11.84PGHH294 pKa = 6.35PPPEE298 pKa = 4.87RR299 pKa = 11.84GSGPRR304 pKa = 11.84GTRR307 pKa = 11.84PPAPSSRR314 pKa = 11.84TGQRR318 pKa = 11.84TRR320 pKa = 11.84QRR322 pKa = 11.84PGHH325 pKa = 6.35PPPEE329 pKa = 4.87RR330 pKa = 11.84GSGPRR335 pKa = 11.84GTRR338 pKa = 11.84PPAPSSRR345 pKa = 11.84TGQRR349 pKa = 11.84TRR351 pKa = 11.84QRR353 pKa = 11.84PGHH356 pKa = 6.35PPPEE360 pKa = 4.87RR361 pKa = 11.84GSGPRR366 pKa = 11.84GTRR369 pKa = 11.84PPAPSSRR376 pKa = 11.84TGQRR380 pKa = 11.84TRR382 pKa = 11.84QRR384 pKa = 11.84PGHH387 pKa = 6.35PPPEE391 pKa = 4.87RR392 pKa = 11.84GSGPRR397 pKa = 11.84GTRR400 pKa = 11.84PPAPSSRR407 pKa = 11.84TGQRR411 pKa = 11.84TRR413 pKa = 11.84QRR415 pKa = 11.84PGHH418 pKa = 6.35PPPEE422 pKa = 4.87RR423 pKa = 11.84GSGPRR428 pKa = 11.84GTRR431 pKa = 11.84PPAPSSRR438 pKa = 11.84TGQRR442 pKa = 11.84TRR444 pKa = 11.84QRR446 pKa = 11.84PGHH449 pKa = 6.35PPPEE453 pKa = 4.87RR454 pKa = 11.84GSGPRR459 pKa = 11.84GTRR462 pKa = 11.84PPAPSSRR469 pKa = 11.84TGQRR473 pKa = 11.84TRR475 pKa = 11.84QRR477 pKa = 11.84PGHH480 pKa = 6.35PPPEE484 pKa = 4.87RR485 pKa = 11.84GSGPRR490 pKa = 11.84GTRR493 pKa = 11.84PPAPSSRR500 pKa = 11.84TGQRR504 pKa = 11.84TRR506 pKa = 11.84QRR508 pKa = 11.84PGHH511 pKa = 6.35PPPEE515 pKa = 4.87RR516 pKa = 11.84GSGPRR521 pKa = 11.84GTRR524 pKa = 11.84PPAPSSRR531 pKa = 11.84TGQRR535 pKa = 11.84TRR537 pKa = 11.84QRR539 pKa = 11.84PGHH542 pKa = 6.35PPPEE546 pKa = 4.87RR547 pKa = 11.84GSGPRR552 pKa = 11.84GTRR555 pKa = 11.84PPAPSSRR562 pKa = 11.84TGQRR566 pKa = 11.84TRR568 pKa = 11.84QRR570 pKa = 11.84PGHH573 pKa = 6.35PPPEE577 pKa = 4.87RR578 pKa = 11.84GSGPRR583 pKa = 11.84GTRR586 pKa = 11.84PPAPSSRR593 pKa = 11.84TGQRR597 pKa = 11.84TRR599 pKa = 11.84QRR601 pKa = 11.84PGHH604 pKa = 6.35PPPEE608 pKa = 4.87RR609 pKa = 11.84GSGPRR614 pKa = 11.84GTRR617 pKa = 11.84PPAPSSRR624 pKa = 11.84TGQRR628 pKa = 11.84TRR630 pKa = 11.84QRR632 pKa = 11.84PGHH635 pKa = 6.35PPPEE639 pKa = 4.87RR640 pKa = 11.84GSGPRR645 pKa = 11.84GTRR648 pKa = 11.84PPAPSSRR655 pKa = 11.84TGQRR659 pKa = 11.84TRR661 pKa = 11.84QRR663 pKa = 11.84PGHH666 pKa = 6.35PPPEE670 pKa = 4.87RR671 pKa = 11.84GSGPRR676 pKa = 11.84GTRR679 pKa = 11.84PPAPSSRR686 pKa = 11.84TGQRR690 pKa = 11.84TRR692 pKa = 11.84QRR694 pKa = 11.84PGHH697 pKa = 6.35PPPEE701 pKa = 4.87RR702 pKa = 11.84GSGPRR707 pKa = 11.84GTRR710 pKa = 11.84PPAPSSRR717 pKa = 11.84TGQRR721 pKa = 11.84TRR723 pKa = 11.84QRR725 pKa = 11.84PGHH728 pKa = 6.35PPPEE732 pKa = 4.87RR733 pKa = 11.84GSGPRR738 pKa = 11.84GTRR741 pKa = 11.84PPAPSSRR748 pKa = 11.84TGQRR752 pKa = 11.84TRR754 pKa = 11.84QRR756 pKa = 11.84PGHH759 pKa = 6.35PPPEE763 pKa = 4.87RR764 pKa = 11.84GSGPRR769 pKa = 11.84GTRR772 pKa = 11.84PPAPSSRR779 pKa = 11.84TGQRR783 pKa = 11.84TRR785 pKa = 11.84QRR787 pKa = 11.84PGHH790 pKa = 6.35PPPEE794 pKa = 4.87RR795 pKa = 11.84GSGPRR800 pKa = 11.84GTRR803 pKa = 11.84PPAPSSRR810 pKa = 11.84TGQRR814 pKa = 11.84TRR816 pKa = 11.84QRR818 pKa = 11.84PGHH821 pKa = 6.35PPPEE825 pKa = 4.87RR826 pKa = 11.84GSGPRR831 pKa = 11.84GTRR834 pKa = 11.84PPAPSSRR841 pKa = 11.84TGQRR845 pKa = 11.84TRR847 pKa = 11.84QRR849 pKa = 11.84PGHH852 pKa = 6.51PPPEE856 pKa = 4.08QTQTLGKK863 pKa = 10.12RR864 pKa = 11.84GARR867 pKa = 11.84VFNNKK872 pKa = 8.0VYY874 pKa = 10.86KK875 pKa = 9.38LTSRR879 pKa = 11.84VCGACGVGVVGG890 pKa = 5.45
MM1 pKa = 7.48GLGFRR6 pKa = 11.84GWGLHH11 pKa = 6.25EE12 pKa = 4.45PACLPGEE19 pKa = 4.31PNQRR23 pKa = 11.84TQGHH27 pKa = 6.82PAPGPSSRR35 pKa = 11.84TGQRR39 pKa = 11.84TRR41 pKa = 11.84QRR43 pKa = 11.84PGHH46 pKa = 6.35PPPEE50 pKa = 4.87RR51 pKa = 11.84GSGPRR56 pKa = 11.84GTRR59 pKa = 11.84PPAPSSRR66 pKa = 11.84TGQRR70 pKa = 11.84TRR72 pKa = 11.84QRR74 pKa = 11.84PGHH77 pKa = 6.35PPPEE81 pKa = 4.87RR82 pKa = 11.84GSGPRR87 pKa = 11.84GTRR90 pKa = 11.84PPAPSSRR97 pKa = 11.84TGQRR101 pKa = 11.84TRR103 pKa = 11.84QRR105 pKa = 11.84PGHH108 pKa = 6.35PPPEE112 pKa = 4.87RR113 pKa = 11.84GSGPRR118 pKa = 11.84GTRR121 pKa = 11.84PPAPSSRR128 pKa = 11.84TGQRR132 pKa = 11.84TRR134 pKa = 11.84QRR136 pKa = 11.84PGHH139 pKa = 6.35PPPEE143 pKa = 4.87RR144 pKa = 11.84GSGPRR149 pKa = 11.84GTRR152 pKa = 11.84PPAPSSRR159 pKa = 11.84TGQRR163 pKa = 11.84TRR165 pKa = 11.84QRR167 pKa = 11.84PGHH170 pKa = 6.35PPPEE174 pKa = 4.87RR175 pKa = 11.84GSGPRR180 pKa = 11.84GTRR183 pKa = 11.84PPAPSSRR190 pKa = 11.84TGQRR194 pKa = 11.84TRR196 pKa = 11.84QRR198 pKa = 11.84PGHH201 pKa = 6.35PPPEE205 pKa = 4.87RR206 pKa = 11.84GSGPRR211 pKa = 11.84GTRR214 pKa = 11.84PPAPSSRR221 pKa = 11.84TGQRR225 pKa = 11.84TRR227 pKa = 11.84QRR229 pKa = 11.84PGHH232 pKa = 6.35PPPEE236 pKa = 4.87RR237 pKa = 11.84GSGPRR242 pKa = 11.84GTRR245 pKa = 11.84PPAPSSRR252 pKa = 11.84TGQRR256 pKa = 11.84TRR258 pKa = 11.84QRR260 pKa = 11.84PGHH263 pKa = 6.35PPPEE267 pKa = 4.87RR268 pKa = 11.84GSGPRR273 pKa = 11.84GTRR276 pKa = 11.84PPAPSSRR283 pKa = 11.84TGQRR287 pKa = 11.84TRR289 pKa = 11.84QRR291 pKa = 11.84PGHH294 pKa = 6.35PPPEE298 pKa = 4.87RR299 pKa = 11.84GSGPRR304 pKa = 11.84GTRR307 pKa = 11.84PPAPSSRR314 pKa = 11.84TGQRR318 pKa = 11.84TRR320 pKa = 11.84QRR322 pKa = 11.84PGHH325 pKa = 6.35PPPEE329 pKa = 4.87RR330 pKa = 11.84GSGPRR335 pKa = 11.84GTRR338 pKa = 11.84PPAPSSRR345 pKa = 11.84TGQRR349 pKa = 11.84TRR351 pKa = 11.84QRR353 pKa = 11.84PGHH356 pKa = 6.35PPPEE360 pKa = 4.87RR361 pKa = 11.84GSGPRR366 pKa = 11.84GTRR369 pKa = 11.84PPAPSSRR376 pKa = 11.84TGQRR380 pKa = 11.84TRR382 pKa = 11.84QRR384 pKa = 11.84PGHH387 pKa = 6.35PPPEE391 pKa = 4.87RR392 pKa = 11.84GSGPRR397 pKa = 11.84GTRR400 pKa = 11.84PPAPSSRR407 pKa = 11.84TGQRR411 pKa = 11.84TRR413 pKa = 11.84QRR415 pKa = 11.84PGHH418 pKa = 6.35PPPEE422 pKa = 4.87RR423 pKa = 11.84GSGPRR428 pKa = 11.84GTRR431 pKa = 11.84PPAPSSRR438 pKa = 11.84TGQRR442 pKa = 11.84TRR444 pKa = 11.84QRR446 pKa = 11.84PGHH449 pKa = 6.35PPPEE453 pKa = 4.87RR454 pKa = 11.84GSGPRR459 pKa = 11.84GTRR462 pKa = 11.84PPAPSSRR469 pKa = 11.84TGQRR473 pKa = 11.84TRR475 pKa = 11.84QRR477 pKa = 11.84PGHH480 pKa = 6.35PPPEE484 pKa = 4.87RR485 pKa = 11.84GSGPRR490 pKa = 11.84GTRR493 pKa = 11.84PPAPSSRR500 pKa = 11.84TGQRR504 pKa = 11.84TRR506 pKa = 11.84QRR508 pKa = 11.84PGHH511 pKa = 6.35PPPEE515 pKa = 4.87RR516 pKa = 11.84GSGPRR521 pKa = 11.84GTRR524 pKa = 11.84PPAPSSRR531 pKa = 11.84TGQRR535 pKa = 11.84TRR537 pKa = 11.84QRR539 pKa = 11.84PGHH542 pKa = 6.35PPPEE546 pKa = 4.87RR547 pKa = 11.84GSGPRR552 pKa = 11.84GTRR555 pKa = 11.84PPAPSSRR562 pKa = 11.84TGQRR566 pKa = 11.84TRR568 pKa = 11.84QRR570 pKa = 11.84PGHH573 pKa = 6.35PPPEE577 pKa = 4.87RR578 pKa = 11.84GSGPRR583 pKa = 11.84GTRR586 pKa = 11.84PPAPSSRR593 pKa = 11.84TGQRR597 pKa = 11.84TRR599 pKa = 11.84QRR601 pKa = 11.84PGHH604 pKa = 6.35PPPEE608 pKa = 4.87RR609 pKa = 11.84GSGPRR614 pKa = 11.84GTRR617 pKa = 11.84PPAPSSRR624 pKa = 11.84TGQRR628 pKa = 11.84TRR630 pKa = 11.84QRR632 pKa = 11.84PGHH635 pKa = 6.35PPPEE639 pKa = 4.87RR640 pKa = 11.84GSGPRR645 pKa = 11.84GTRR648 pKa = 11.84PPAPSSRR655 pKa = 11.84TGQRR659 pKa = 11.84TRR661 pKa = 11.84QRR663 pKa = 11.84PGHH666 pKa = 6.35PPPEE670 pKa = 4.87RR671 pKa = 11.84GSGPRR676 pKa = 11.84GTRR679 pKa = 11.84PPAPSSRR686 pKa = 11.84TGQRR690 pKa = 11.84TRR692 pKa = 11.84QRR694 pKa = 11.84PGHH697 pKa = 6.35PPPEE701 pKa = 4.87RR702 pKa = 11.84GSGPRR707 pKa = 11.84GTRR710 pKa = 11.84PPAPSSRR717 pKa = 11.84TGQRR721 pKa = 11.84TRR723 pKa = 11.84QRR725 pKa = 11.84PGHH728 pKa = 6.35PPPEE732 pKa = 4.87RR733 pKa = 11.84GSGPRR738 pKa = 11.84GTRR741 pKa = 11.84PPAPSSRR748 pKa = 11.84TGQRR752 pKa = 11.84TRR754 pKa = 11.84QRR756 pKa = 11.84PGHH759 pKa = 6.35PPPEE763 pKa = 4.87RR764 pKa = 11.84GSGPRR769 pKa = 11.84GTRR772 pKa = 11.84PPAPSSRR779 pKa = 11.84TGQRR783 pKa = 11.84TRR785 pKa = 11.84QRR787 pKa = 11.84PGHH790 pKa = 6.35PPPEE794 pKa = 4.87RR795 pKa = 11.84GSGPRR800 pKa = 11.84GTRR803 pKa = 11.84PPAPSSRR810 pKa = 11.84TGQRR814 pKa = 11.84TRR816 pKa = 11.84QRR818 pKa = 11.84PGHH821 pKa = 6.35PPPEE825 pKa = 4.87RR826 pKa = 11.84GSGPRR831 pKa = 11.84GTRR834 pKa = 11.84PPAPSSRR841 pKa = 11.84TGQRR845 pKa = 11.84TRR847 pKa = 11.84QRR849 pKa = 11.84PGHH852 pKa = 6.51PPPEE856 pKa = 4.08QTQTLGKK863 pKa = 10.12RR864 pKa = 11.84GARR867 pKa = 11.84VFNNKK872 pKa = 8.0VYY874 pKa = 10.86KK875 pKa = 9.38LTSRR879 pKa = 11.84VCGACGVGVVGG890 pKa = 5.45
Molecular weight: 95.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
40829 |
59 |
3105 |
510.4 |
55.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.452 ± 0.461 | 1.876 ± 0.164 |
4.257 ± 0.162 | 5.643 ± 0.208 |
3.644 ± 0.206 | 7.781 ± 0.46 |
2.459 ± 0.115 | 3.331 ± 0.202 |
2.959 ± 0.191 | 10.216 ± 0.361 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.842 ± 0.116 | 2.851 ± 0.224 |
8.758 ± 0.64 | 3.975 ± 0.202 |
7.196 ± 0.382 | 7.272 ± 0.18 |
6.03 ± 0.262 | 6.486 ± 0.233 |
1.203 ± 0.082 | 2.77 ± 0.145 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
