
bacterium D16-63
Taxonomy: cellular organisms; Bacteria; unclassified Bacteria
Average proteome isoelectric point is 5.73
Get precalculated fractions of proteins
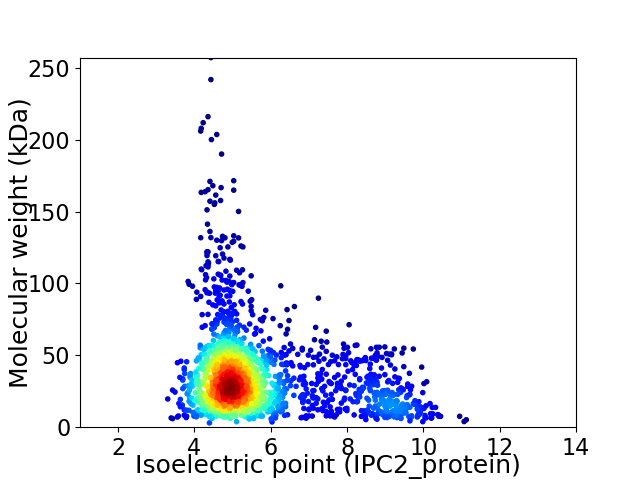
Virtual 2D-PAGE plot for 1845 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3A9AVH4|A0A3A9AVH4_9BACT ATP-binding protein OS=bacterium D16-63 OX=2320119 GN=D7X10_08945 PE=4 SV=1
MM1 pKa = 7.76RR2 pKa = 11.84APVVSRR8 pKa = 11.84TSKK11 pKa = 10.66LSAVLLVLLLACTLGPAAMADD32 pKa = 3.81EE33 pKa = 4.97PSADD37 pKa = 3.6SAVGGVGADD46 pKa = 3.21AADD49 pKa = 3.89AEE51 pKa = 4.71GAPMLASLMASGAGHH66 pKa = 7.43ISTAVALEE74 pKa = 4.59DD75 pKa = 4.03DD76 pKa = 4.58PEE78 pKa = 4.29PVVGSFTVGGLTYY91 pKa = 10.43AIVSEE96 pKa = 4.69GEE98 pKa = 4.21VVLVSVGPDD107 pKa = 2.9ADD109 pKa = 3.73ALAVGSNGAGEE120 pKa = 4.33GLLANSSGAPSGEE133 pKa = 4.14DD134 pKa = 2.91SGSGVPPRR142 pKa = 11.84SVAEE146 pKa = 4.0QVPSGAASLSPSPEE160 pKa = 3.81GVPSDD165 pKa = 3.58GAEE168 pKa = 4.38GEE170 pKa = 4.51DD171 pKa = 3.65ADD173 pKa = 4.72DD174 pKa = 4.18PVVLEE179 pKa = 4.26VPEE182 pKa = 4.24SVEE185 pKa = 4.03FDD187 pKa = 3.08GVAYY191 pKa = 10.17SVVAIGPRR199 pKa = 11.84ALAGCDD205 pKa = 2.87ADD207 pKa = 5.43AIVIPTTVGSVDD219 pKa = 3.16EE220 pKa = 4.19LAFRR224 pKa = 11.84GSAVKK229 pKa = 10.45AIEE232 pKa = 4.11VAGGNASFSSFDD244 pKa = 3.17GMLFDD249 pKa = 5.53AEE251 pKa = 4.69RR252 pKa = 11.84ISLLLIPEE260 pKa = 5.1GKK262 pKa = 9.87QGAARR267 pKa = 11.84IPSTAEE273 pKa = 3.49AVPPGAFSHH282 pKa = 6.3CASVTSLSVDD292 pKa = 3.26AGSAAYY298 pKa = 10.44LSRR301 pKa = 11.84NGCLYY306 pKa = 10.56DD307 pKa = 3.66ASGEE311 pKa = 4.01ALLRR315 pKa = 11.84VPAGATEE322 pKa = 3.89IVIADD327 pKa = 3.93GCTAVAAGALEE338 pKa = 4.62GCAKK342 pKa = 10.16LRR344 pKa = 11.84AIQASASVTEE354 pKa = 4.02VSPDD358 pKa = 3.36VLGAAVAADD367 pKa = 3.89GGSAPVASLGSSGALAADD385 pKa = 4.98GGSQMGDD392 pKa = 2.97VGDD395 pKa = 4.34ADD397 pKa = 3.73AAEE400 pKa = 5.03GGSADD405 pKa = 4.07GPQAPFGDD413 pKa = 4.08EE414 pKa = 4.15GRR416 pKa = 11.84GLDD419 pKa = 4.47AEE421 pKa = 4.6AEE423 pKa = 4.29APAGSSPAVSSQLTSLVVLATAGDD447 pKa = 4.43DD448 pKa = 3.8LPQVNSTSIIVSLPEE463 pKa = 4.27GADD466 pKa = 3.48PAPWEE471 pKa = 3.93AVGFEE476 pKa = 4.41VEE478 pKa = 5.07VIAADD483 pKa = 3.5ADD485 pKa = 4.19GNAEE489 pKa = 4.17TVEE492 pKa = 4.28GEE494 pKa = 4.32SLDD497 pKa = 5.54DD498 pKa = 3.53ITPEE502 pKa = 4.21DD503 pKa = 3.79AHH505 pKa = 7.81DD506 pKa = 4.47HH507 pKa = 7.08DD508 pKa = 5.35VDD510 pKa = 4.25EE511 pKa = 5.75PDD513 pKa = 4.84DD514 pKa = 4.52APSDD518 pKa = 3.88GPVIVDD524 pKa = 4.19EE525 pKa = 4.23VTIDD529 pKa = 3.24YY530 pKa = 10.48GYY532 pKa = 11.4ANEE535 pKa = 4.49RR536 pKa = 11.84SSADD540 pKa = 3.12GAYY543 pKa = 9.59EE544 pKa = 3.82RR545 pKa = 11.84AVPKK549 pKa = 10.16IDD551 pKa = 4.9AVLKK555 pKa = 8.68LQPITAEE562 pKa = 3.73EE563 pKa = 4.09LMEE566 pKa = 4.5GEE568 pKa = 4.23VAARR572 pKa = 11.84GEE574 pKa = 4.16DD575 pKa = 3.42TSAGFGSAADD585 pKa = 4.23GSADD589 pKa = 3.86GACGAVDD596 pKa = 3.7SDD598 pKa = 3.8AGSGGGAAADD608 pKa = 3.86AADD611 pKa = 4.23ADD613 pKa = 4.21EE614 pKa = 5.33GEE616 pKa = 4.35PTLASKK622 pKa = 10.64LADD625 pKa = 3.77LPEE628 pKa = 5.11DD629 pKa = 3.83SQATASFLYY638 pKa = 9.9EE639 pKa = 3.82GCLYY643 pKa = 10.99VIDD646 pKa = 4.71PEE648 pKa = 5.38GGAALVAVDD657 pKa = 4.37PKK659 pKa = 10.87RR660 pKa = 11.84LAEE663 pKa = 4.19CVEE666 pKa = 4.6DD667 pKa = 3.93PTTLILPDD675 pKa = 3.89YY676 pKa = 10.42VDD678 pKa = 4.86DD679 pKa = 4.68GTWSYY684 pKa = 11.5PLTRR688 pKa = 11.84IAKK691 pKa = 8.7GALRR695 pKa = 11.84GSGIEE700 pKa = 4.11RR701 pKa = 11.84LWIPASATYY710 pKa = 10.67LDD712 pKa = 4.29YY713 pKa = 11.35ALEE716 pKa = 4.51GCEE719 pKa = 3.57TLRR722 pKa = 11.84YY723 pKa = 10.06VEE725 pKa = 4.45ISEE728 pKa = 5.15DD729 pKa = 3.58SPAYY733 pKa = 9.88SSKK736 pKa = 10.92GGCLYY741 pKa = 11.0DD742 pKa = 4.01KK743 pKa = 10.91AGEE746 pKa = 4.2KK747 pKa = 10.38LWMVPEE753 pKa = 4.65GGGDD757 pKa = 3.19IAIEE761 pKa = 4.07VAVDD765 pKa = 3.75GGAGEE770 pKa = 4.51INPARR775 pKa = 11.84QTDD778 pKa = 3.72QAKK781 pKa = 8.66LTNIAQMHH789 pKa = 5.67YY790 pKa = 9.61PASLSNLSTARR801 pKa = 11.84APRR804 pKa = 11.84TSYY807 pKa = 9.44RR808 pKa = 11.84TTHH811 pKa = 7.06LYY813 pKa = 6.35TTNFHH818 pKa = 6.74SVTDD822 pKa = 3.68YY823 pKa = 7.64TTQVDD828 pKa = 3.43VDD830 pKa = 4.16GTCVSGCEE838 pKa = 4.09TDD840 pKa = 4.08DD841 pKa = 5.04FPWTLIWTLRR851 pKa = 11.84RR852 pKa = 11.84QITNDD857 pKa = 2.91RR858 pKa = 11.84GIYY861 pKa = 9.97FYY863 pKa = 10.88TDD865 pKa = 3.22GTISEE870 pKa = 4.35MDD872 pKa = 3.57PYY874 pKa = 11.17GQLDD878 pKa = 3.77YY879 pKa = 11.26QYY881 pKa = 10.8VNTISSSRR889 pKa = 11.84IYY891 pKa = 9.39RR892 pKa = 11.84THH894 pKa = 6.4LAAWSSSPLGPPLTHH909 pKa = 6.73SLHH912 pKa = 6.18TGPIYY917 pKa = 9.6LHH919 pKa = 6.82CGVGYY924 pKa = 10.57LKK926 pKa = 9.33QTVTWADD933 pKa = 3.33TATGSTQTEE942 pKa = 4.24TVPLNGATTAPSPSRR957 pKa = 11.84TGYY960 pKa = 8.41TFAGWYY966 pKa = 5.89TAASGGSYY974 pKa = 10.43VCGAGGSVTVTSDD987 pKa = 3.0VTYY990 pKa = 9.81YY991 pKa = 11.28AHH993 pKa = 6.65WTPNTYY999 pKa = 8.66TVSS1002 pKa = 3.29
MM1 pKa = 7.76RR2 pKa = 11.84APVVSRR8 pKa = 11.84TSKK11 pKa = 10.66LSAVLLVLLLACTLGPAAMADD32 pKa = 3.81EE33 pKa = 4.97PSADD37 pKa = 3.6SAVGGVGADD46 pKa = 3.21AADD49 pKa = 3.89AEE51 pKa = 4.71GAPMLASLMASGAGHH66 pKa = 7.43ISTAVALEE74 pKa = 4.59DD75 pKa = 4.03DD76 pKa = 4.58PEE78 pKa = 4.29PVVGSFTVGGLTYY91 pKa = 10.43AIVSEE96 pKa = 4.69GEE98 pKa = 4.21VVLVSVGPDD107 pKa = 2.9ADD109 pKa = 3.73ALAVGSNGAGEE120 pKa = 4.33GLLANSSGAPSGEE133 pKa = 4.14DD134 pKa = 2.91SGSGVPPRR142 pKa = 11.84SVAEE146 pKa = 4.0QVPSGAASLSPSPEE160 pKa = 3.81GVPSDD165 pKa = 3.58GAEE168 pKa = 4.38GEE170 pKa = 4.51DD171 pKa = 3.65ADD173 pKa = 4.72DD174 pKa = 4.18PVVLEE179 pKa = 4.26VPEE182 pKa = 4.24SVEE185 pKa = 4.03FDD187 pKa = 3.08GVAYY191 pKa = 10.17SVVAIGPRR199 pKa = 11.84ALAGCDD205 pKa = 2.87ADD207 pKa = 5.43AIVIPTTVGSVDD219 pKa = 3.16EE220 pKa = 4.19LAFRR224 pKa = 11.84GSAVKK229 pKa = 10.45AIEE232 pKa = 4.11VAGGNASFSSFDD244 pKa = 3.17GMLFDD249 pKa = 5.53AEE251 pKa = 4.69RR252 pKa = 11.84ISLLLIPEE260 pKa = 5.1GKK262 pKa = 9.87QGAARR267 pKa = 11.84IPSTAEE273 pKa = 3.49AVPPGAFSHH282 pKa = 6.3CASVTSLSVDD292 pKa = 3.26AGSAAYY298 pKa = 10.44LSRR301 pKa = 11.84NGCLYY306 pKa = 10.56DD307 pKa = 3.66ASGEE311 pKa = 4.01ALLRR315 pKa = 11.84VPAGATEE322 pKa = 3.89IVIADD327 pKa = 3.93GCTAVAAGALEE338 pKa = 4.62GCAKK342 pKa = 10.16LRR344 pKa = 11.84AIQASASVTEE354 pKa = 4.02VSPDD358 pKa = 3.36VLGAAVAADD367 pKa = 3.89GGSAPVASLGSSGALAADD385 pKa = 4.98GGSQMGDD392 pKa = 2.97VGDD395 pKa = 4.34ADD397 pKa = 3.73AAEE400 pKa = 5.03GGSADD405 pKa = 4.07GPQAPFGDD413 pKa = 4.08EE414 pKa = 4.15GRR416 pKa = 11.84GLDD419 pKa = 4.47AEE421 pKa = 4.6AEE423 pKa = 4.29APAGSSPAVSSQLTSLVVLATAGDD447 pKa = 4.43DD448 pKa = 3.8LPQVNSTSIIVSLPEE463 pKa = 4.27GADD466 pKa = 3.48PAPWEE471 pKa = 3.93AVGFEE476 pKa = 4.41VEE478 pKa = 5.07VIAADD483 pKa = 3.5ADD485 pKa = 4.19GNAEE489 pKa = 4.17TVEE492 pKa = 4.28GEE494 pKa = 4.32SLDD497 pKa = 5.54DD498 pKa = 3.53ITPEE502 pKa = 4.21DD503 pKa = 3.79AHH505 pKa = 7.81DD506 pKa = 4.47HH507 pKa = 7.08DD508 pKa = 5.35VDD510 pKa = 4.25EE511 pKa = 5.75PDD513 pKa = 4.84DD514 pKa = 4.52APSDD518 pKa = 3.88GPVIVDD524 pKa = 4.19EE525 pKa = 4.23VTIDD529 pKa = 3.24YY530 pKa = 10.48GYY532 pKa = 11.4ANEE535 pKa = 4.49RR536 pKa = 11.84SSADD540 pKa = 3.12GAYY543 pKa = 9.59EE544 pKa = 3.82RR545 pKa = 11.84AVPKK549 pKa = 10.16IDD551 pKa = 4.9AVLKK555 pKa = 8.68LQPITAEE562 pKa = 3.73EE563 pKa = 4.09LMEE566 pKa = 4.5GEE568 pKa = 4.23VAARR572 pKa = 11.84GEE574 pKa = 4.16DD575 pKa = 3.42TSAGFGSAADD585 pKa = 4.23GSADD589 pKa = 3.86GACGAVDD596 pKa = 3.7SDD598 pKa = 3.8AGSGGGAAADD608 pKa = 3.86AADD611 pKa = 4.23ADD613 pKa = 4.21EE614 pKa = 5.33GEE616 pKa = 4.35PTLASKK622 pKa = 10.64LADD625 pKa = 3.77LPEE628 pKa = 5.11DD629 pKa = 3.83SQATASFLYY638 pKa = 9.9EE639 pKa = 3.82GCLYY643 pKa = 10.99VIDD646 pKa = 4.71PEE648 pKa = 5.38GGAALVAVDD657 pKa = 4.37PKK659 pKa = 10.87RR660 pKa = 11.84LAEE663 pKa = 4.19CVEE666 pKa = 4.6DD667 pKa = 3.93PTTLILPDD675 pKa = 3.89YY676 pKa = 10.42VDD678 pKa = 4.86DD679 pKa = 4.68GTWSYY684 pKa = 11.5PLTRR688 pKa = 11.84IAKK691 pKa = 8.7GALRR695 pKa = 11.84GSGIEE700 pKa = 4.11RR701 pKa = 11.84LWIPASATYY710 pKa = 10.67LDD712 pKa = 4.29YY713 pKa = 11.35ALEE716 pKa = 4.51GCEE719 pKa = 3.57TLRR722 pKa = 11.84YY723 pKa = 10.06VEE725 pKa = 4.45ISEE728 pKa = 5.15DD729 pKa = 3.58SPAYY733 pKa = 9.88SSKK736 pKa = 10.92GGCLYY741 pKa = 11.0DD742 pKa = 4.01KK743 pKa = 10.91AGEE746 pKa = 4.2KK747 pKa = 10.38LWMVPEE753 pKa = 4.65GGGDD757 pKa = 3.19IAIEE761 pKa = 4.07VAVDD765 pKa = 3.75GGAGEE770 pKa = 4.51INPARR775 pKa = 11.84QTDD778 pKa = 3.72QAKK781 pKa = 8.66LTNIAQMHH789 pKa = 5.67YY790 pKa = 9.61PASLSNLSTARR801 pKa = 11.84APRR804 pKa = 11.84TSYY807 pKa = 9.44RR808 pKa = 11.84TTHH811 pKa = 7.06LYY813 pKa = 6.35TTNFHH818 pKa = 6.74SVTDD822 pKa = 3.68YY823 pKa = 7.64TTQVDD828 pKa = 3.43VDD830 pKa = 4.16GTCVSGCEE838 pKa = 4.09TDD840 pKa = 4.08DD841 pKa = 5.04FPWTLIWTLRR851 pKa = 11.84RR852 pKa = 11.84QITNDD857 pKa = 2.91RR858 pKa = 11.84GIYY861 pKa = 9.97FYY863 pKa = 10.88TDD865 pKa = 3.22GTISEE870 pKa = 4.35MDD872 pKa = 3.57PYY874 pKa = 11.17GQLDD878 pKa = 3.77YY879 pKa = 11.26QYY881 pKa = 10.8VNTISSSRR889 pKa = 11.84IYY891 pKa = 9.39RR892 pKa = 11.84THH894 pKa = 6.4LAAWSSSPLGPPLTHH909 pKa = 6.73SLHH912 pKa = 6.18TGPIYY917 pKa = 9.6LHH919 pKa = 6.82CGVGYY924 pKa = 10.57LKK926 pKa = 9.33QTVTWADD933 pKa = 3.33TATGSTQTEE942 pKa = 4.24TVPLNGATTAPSPSRR957 pKa = 11.84TGYY960 pKa = 8.41TFAGWYY966 pKa = 5.89TAASGGSYY974 pKa = 10.43VCGAGGSVTVTSDD987 pKa = 3.0VTYY990 pKa = 9.81YY991 pKa = 11.28AHH993 pKa = 6.65WTPNTYY999 pKa = 8.66TVSS1002 pKa = 3.29
Molecular weight: 101.42 kDa
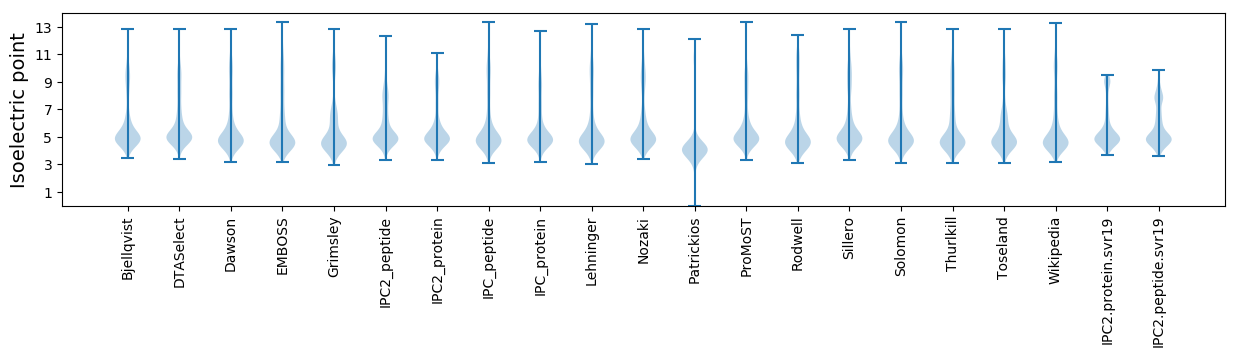
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3A9B2R6|A0A3A9B2R6_9BACT Alpha/beta hydrolase OS=bacterium D16-63 OX=2320119 GN=D7X10_07525 PE=4 SV=1
GG1 pKa = 7.38RR2 pKa = 11.84GGPGGRR8 pKa = 11.84GGPGAGRR15 pKa = 11.84GAGGRR20 pKa = 11.84GAAHH24 pKa = 6.17QGGGQGPGRR33 pKa = 11.84GVRR36 pKa = 11.84HH37 pKa = 5.91RR38 pKa = 11.84AQRR41 pKa = 11.84GLL43 pKa = 3.46
GG1 pKa = 7.38RR2 pKa = 11.84GGPGGRR8 pKa = 11.84GGPGAGRR15 pKa = 11.84GAGGRR20 pKa = 11.84GAAHH24 pKa = 6.17QGGGQGPGRR33 pKa = 11.84GVRR36 pKa = 11.84HH37 pKa = 5.91RR38 pKa = 11.84AQRR41 pKa = 11.84GLL43 pKa = 3.46
Molecular weight: 3.93 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
639091 |
27 |
2417 |
346.4 |
37.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.361 ± 0.083 | 1.411 ± 0.027 |
5.943 ± 0.048 | 6.612 ± 0.063 |
3.432 ± 0.038 | 9.161 ± 0.061 |
1.666 ± 0.022 | 4.075 ± 0.051 |
3.136 ± 0.047 | 9.228 ± 0.074 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.44 ± 0.034 | 2.337 ± 0.035 |
4.963 ± 0.041 | 2.891 ± 0.025 |
6.115 ± 0.062 | 5.871 ± 0.07 |
5.2 ± 0.068 | 8.002 ± 0.053 |
1.267 ± 0.034 | 2.891 ± 0.038 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
