
Bark beetle-associated genomovirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemycircularvirus; Gemycircularvirus denbre1
Average proteome isoelectric point is 7.34
Get precalculated fractions of proteins
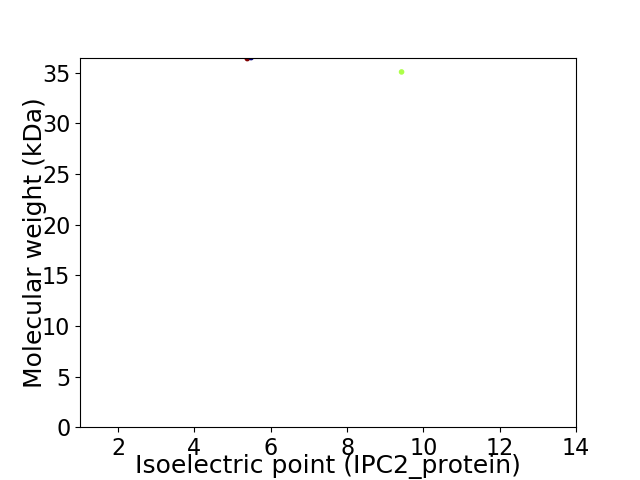
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
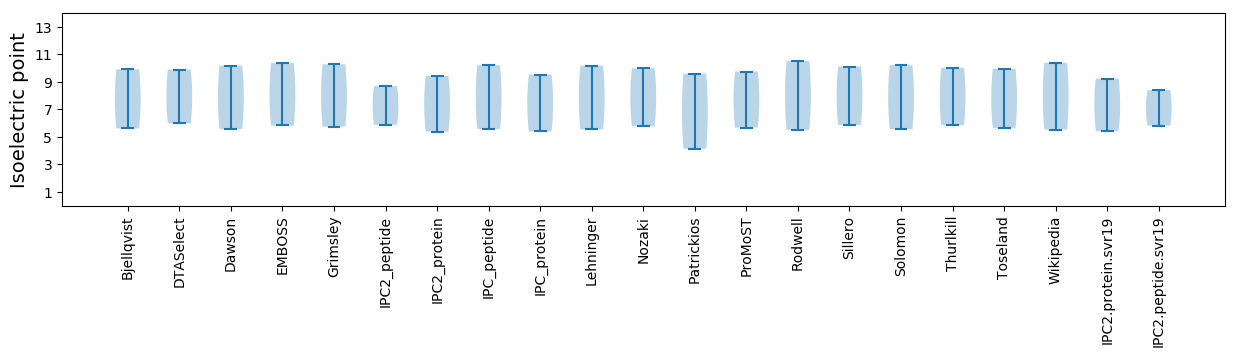
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A344A3P5|A0A344A3P5_9VIRU Replication-associated protein OS=Bark beetle-associated genomovirus 1 OX=2230896 PE=3 SV=1
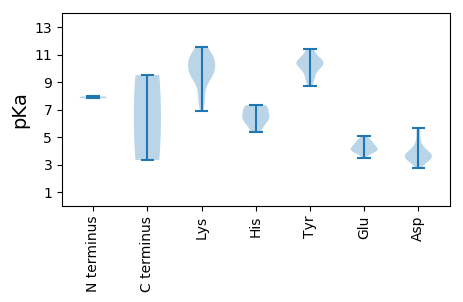
MM1 pKa = 7.82PFACNARR8 pKa = 11.84YY9 pKa = 8.96FLVTYY14 pKa = 8.7GHH16 pKa = 6.38VEE18 pKa = 4.01SLDD21 pKa = 3.42PFALVDD27 pKa = 3.58HH28 pKa = 6.89FGKK31 pKa = 10.67LGAEE35 pKa = 4.21IIVSLEE41 pKa = 3.82QYY43 pKa = 10.33NATLGVHH50 pKa = 5.51FHH52 pKa = 6.04VFADD56 pKa = 4.64FGRR59 pKa = 11.84KK60 pKa = 8.16FRR62 pKa = 11.84SRR64 pKa = 11.84RR65 pKa = 11.84TDD67 pKa = 2.73IFDD70 pKa = 3.67VEE72 pKa = 4.64GFHH75 pKa = 7.31PNISPSRR82 pKa = 11.84GTPEE86 pKa = 3.98AGYY89 pKa = 10.62DD90 pKa = 3.63YY91 pKa = 10.68AIKK94 pKa = 10.92DD95 pKa = 3.59GDD97 pKa = 4.02VVAGGLARR105 pKa = 11.84PSGVVASGRR114 pKa = 11.84QAKK117 pKa = 9.23WNTILDD123 pKa = 3.75AEE125 pKa = 4.57TRR127 pKa = 11.84DD128 pKa = 3.91EE129 pKa = 4.71FCALCEE135 pKa = 3.95EE136 pKa = 5.09LDD138 pKa = 3.97RR139 pKa = 11.84EE140 pKa = 4.14RR141 pKa = 11.84LVCSFGQIQKK151 pKa = 9.89FADD154 pKa = 2.8WRR156 pKa = 11.84FAVEE160 pKa = 3.92PEE162 pKa = 4.75PYY164 pKa = 10.07VSPDD168 pKa = 3.13GVFDD172 pKa = 4.04LADD175 pKa = 4.02YY176 pKa = 11.38GDD178 pKa = 3.67LGYY181 pKa = 10.33WRR183 pKa = 11.84DD184 pKa = 3.43NFLFADD190 pKa = 3.56TTGRR194 pKa = 11.84SKK196 pKa = 11.16SLILWGPSRR205 pKa = 11.84MGKK208 pKa = 6.86TVWARR213 pKa = 11.84SLGRR217 pKa = 11.84HH218 pKa = 6.15LYY220 pKa = 10.37FGGIFSARR228 pKa = 11.84NIGDD232 pKa = 3.2DD233 pKa = 3.86RR234 pKa = 11.84IRR236 pKa = 11.84FAVFDD241 pKa = 5.4DD242 pKa = 3.3IAGGIKK248 pKa = 10.03FFPRR252 pKa = 11.84FKK254 pKa = 10.79DD255 pKa = 3.12WLGCQMEE262 pKa = 4.42FMVKK266 pKa = 9.55EE267 pKa = 4.57LYY269 pKa = 10.07RR270 pKa = 11.84DD271 pKa = 3.33PHH273 pKa = 6.29LFRR276 pKa = 11.84WGRR279 pKa = 11.84PAIWIANSDD288 pKa = 3.56PRR290 pKa = 11.84HH291 pKa = 6.58DD292 pKa = 3.89MTHH295 pKa = 7.25DD296 pKa = 4.74DD297 pKa = 4.35IVWLEE302 pKa = 3.92ANCIFVEE309 pKa = 4.22ISSPIFRR316 pKa = 11.84ANTT319 pKa = 3.33
MM1 pKa = 7.82PFACNARR8 pKa = 11.84YY9 pKa = 8.96FLVTYY14 pKa = 8.7GHH16 pKa = 6.38VEE18 pKa = 4.01SLDD21 pKa = 3.42PFALVDD27 pKa = 3.58HH28 pKa = 6.89FGKK31 pKa = 10.67LGAEE35 pKa = 4.21IIVSLEE41 pKa = 3.82QYY43 pKa = 10.33NATLGVHH50 pKa = 5.51FHH52 pKa = 6.04VFADD56 pKa = 4.64FGRR59 pKa = 11.84KK60 pKa = 8.16FRR62 pKa = 11.84SRR64 pKa = 11.84RR65 pKa = 11.84TDD67 pKa = 2.73IFDD70 pKa = 3.67VEE72 pKa = 4.64GFHH75 pKa = 7.31PNISPSRR82 pKa = 11.84GTPEE86 pKa = 3.98AGYY89 pKa = 10.62DD90 pKa = 3.63YY91 pKa = 10.68AIKK94 pKa = 10.92DD95 pKa = 3.59GDD97 pKa = 4.02VVAGGLARR105 pKa = 11.84PSGVVASGRR114 pKa = 11.84QAKK117 pKa = 9.23WNTILDD123 pKa = 3.75AEE125 pKa = 4.57TRR127 pKa = 11.84DD128 pKa = 3.91EE129 pKa = 4.71FCALCEE135 pKa = 3.95EE136 pKa = 5.09LDD138 pKa = 3.97RR139 pKa = 11.84EE140 pKa = 4.14RR141 pKa = 11.84LVCSFGQIQKK151 pKa = 9.89FADD154 pKa = 2.8WRR156 pKa = 11.84FAVEE160 pKa = 3.92PEE162 pKa = 4.75PYY164 pKa = 10.07VSPDD168 pKa = 3.13GVFDD172 pKa = 4.04LADD175 pKa = 4.02YY176 pKa = 11.38GDD178 pKa = 3.67LGYY181 pKa = 10.33WRR183 pKa = 11.84DD184 pKa = 3.43NFLFADD190 pKa = 3.56TTGRR194 pKa = 11.84SKK196 pKa = 11.16SLILWGPSRR205 pKa = 11.84MGKK208 pKa = 6.86TVWARR213 pKa = 11.84SLGRR217 pKa = 11.84HH218 pKa = 6.15LYY220 pKa = 10.37FGGIFSARR228 pKa = 11.84NIGDD232 pKa = 3.2DD233 pKa = 3.86RR234 pKa = 11.84IRR236 pKa = 11.84FAVFDD241 pKa = 5.4DD242 pKa = 3.3IAGGIKK248 pKa = 10.03FFPRR252 pKa = 11.84FKK254 pKa = 10.79DD255 pKa = 3.12WLGCQMEE262 pKa = 4.42FMVKK266 pKa = 9.55EE267 pKa = 4.57LYY269 pKa = 10.07RR270 pKa = 11.84DD271 pKa = 3.33PHH273 pKa = 6.29LFRR276 pKa = 11.84WGRR279 pKa = 11.84PAIWIANSDD288 pKa = 3.56PRR290 pKa = 11.84HH291 pKa = 6.58DD292 pKa = 3.89MTHH295 pKa = 7.25DD296 pKa = 4.74DD297 pKa = 4.35IVWLEE302 pKa = 3.92ANCIFVEE309 pKa = 4.22ISSPIFRR316 pKa = 11.84ANTT319 pKa = 3.33
Molecular weight: 36.37 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A344A3P5|A0A344A3P5_9VIRU Replication-associated protein OS=Bark beetle-associated genomovirus 1 OX=2230896 PE=3 SV=1
MM1 pKa = 7.95AYY3 pKa = 10.09RR4 pKa = 11.84RR5 pKa = 11.84GGTARR10 pKa = 11.84KK11 pKa = 9.52SYY13 pKa = 10.48KK14 pKa = 9.14ATAKK18 pKa = 9.79RR19 pKa = 11.84RR20 pKa = 11.84YY21 pKa = 9.36AGRR24 pKa = 11.84SKK26 pKa = 10.51FRR28 pKa = 11.84RR29 pKa = 11.84SYY31 pKa = 10.51AKK33 pKa = 9.91KK34 pKa = 7.74RR35 pKa = 11.84TYY37 pKa = 10.27RR38 pKa = 11.84KK39 pKa = 9.67RR40 pKa = 11.84LISIKK45 pKa = 10.51AVLNVTSRR53 pKa = 11.84KK54 pKa = 9.23KK55 pKa = 10.47RR56 pKa = 11.84NGMLSWSNTNPGTGASQTIAAGGAVVAGNSVGFFLFSPTCMNLNQGSSNPTYY108 pKa = 10.4FINSAEE114 pKa = 4.24RR115 pKa = 11.84TSTTCYY121 pKa = 9.46MRR123 pKa = 11.84GFSEE127 pKa = 4.31TLRR130 pKa = 11.84IQTNSHH136 pKa = 5.34VPWFHH141 pKa = 7.26RR142 pKa = 11.84RR143 pKa = 11.84ICFCFRR149 pKa = 11.84GATPFASPNSSDD161 pKa = 3.36TPTQNMSPYY170 pKa = 9.75VDD172 pKa = 3.45TSNGMEE178 pKa = 4.14RR179 pKa = 11.84LWLNQQVNAMGQSVQAIWGVLFKK202 pKa = 11.39GMINQDD208 pKa = 2.97WNDD211 pKa = 3.5VVIAPVDD218 pKa = 3.6TTRR221 pKa = 11.84VDD223 pKa = 3.78LKK225 pKa = 10.96FDD227 pKa = 3.58KK228 pKa = 10.49TWLIKK233 pKa = 10.62SGNEE237 pKa = 3.51SGTIVQRR244 pKa = 11.84KK245 pKa = 7.82LWHH248 pKa = 6.5PMNKK252 pKa = 9.58NLVYY256 pKa = 10.84DD257 pKa = 4.31DD258 pKa = 5.62DD259 pKa = 4.21EE260 pKa = 4.93TGEE263 pKa = 4.4SMASQYY269 pKa = 11.39YY270 pKa = 10.32SVDD273 pKa = 3.43SKK275 pKa = 11.56QGMGDD280 pKa = 3.65FYY282 pKa = 11.23VYY284 pKa = 10.59DD285 pKa = 4.98IITPGLGGSATDD297 pKa = 5.45LISIQANSTMYY308 pKa = 9.35WHH310 pKa = 7.07EE311 pKa = 4.05KK312 pKa = 9.5
MM1 pKa = 7.95AYY3 pKa = 10.09RR4 pKa = 11.84RR5 pKa = 11.84GGTARR10 pKa = 11.84KK11 pKa = 9.52SYY13 pKa = 10.48KK14 pKa = 9.14ATAKK18 pKa = 9.79RR19 pKa = 11.84RR20 pKa = 11.84YY21 pKa = 9.36AGRR24 pKa = 11.84SKK26 pKa = 10.51FRR28 pKa = 11.84RR29 pKa = 11.84SYY31 pKa = 10.51AKK33 pKa = 9.91KK34 pKa = 7.74RR35 pKa = 11.84TYY37 pKa = 10.27RR38 pKa = 11.84KK39 pKa = 9.67RR40 pKa = 11.84LISIKK45 pKa = 10.51AVLNVTSRR53 pKa = 11.84KK54 pKa = 9.23KK55 pKa = 10.47RR56 pKa = 11.84NGMLSWSNTNPGTGASQTIAAGGAVVAGNSVGFFLFSPTCMNLNQGSSNPTYY108 pKa = 10.4FINSAEE114 pKa = 4.24RR115 pKa = 11.84TSTTCYY121 pKa = 9.46MRR123 pKa = 11.84GFSEE127 pKa = 4.31TLRR130 pKa = 11.84IQTNSHH136 pKa = 5.34VPWFHH141 pKa = 7.26RR142 pKa = 11.84RR143 pKa = 11.84ICFCFRR149 pKa = 11.84GATPFASPNSSDD161 pKa = 3.36TPTQNMSPYY170 pKa = 9.75VDD172 pKa = 3.45TSNGMEE178 pKa = 4.14RR179 pKa = 11.84LWLNQQVNAMGQSVQAIWGVLFKK202 pKa = 11.39GMINQDD208 pKa = 2.97WNDD211 pKa = 3.5VVIAPVDD218 pKa = 3.6TTRR221 pKa = 11.84VDD223 pKa = 3.78LKK225 pKa = 10.96FDD227 pKa = 3.58KK228 pKa = 10.49TWLIKK233 pKa = 10.62SGNEE237 pKa = 3.51SGTIVQRR244 pKa = 11.84KK245 pKa = 7.82LWHH248 pKa = 6.5PMNKK252 pKa = 9.58NLVYY256 pKa = 10.84DD257 pKa = 4.31DD258 pKa = 5.62DD259 pKa = 4.21EE260 pKa = 4.93TGEE263 pKa = 4.4SMASQYY269 pKa = 11.39YY270 pKa = 10.32SVDD273 pKa = 3.43SKK275 pKa = 11.56QGMGDD280 pKa = 3.65FYY282 pKa = 11.23VYY284 pKa = 10.59DD285 pKa = 4.98IITPGLGGSATDD297 pKa = 5.45LISIQANSTMYY308 pKa = 9.35WHH310 pKa = 7.07EE311 pKa = 4.05KK312 pKa = 9.5
Molecular weight: 35.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
631 |
312 |
319 |
315.5 |
35.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.448 ± 0.517 | 1.585 ± 0.218 |
6.656 ± 1.561 | 3.645 ± 1.009 |
6.498 ± 1.678 | 8.399 ± 0.278 |
2.06 ± 0.56 | 5.388 ± 0.418 |
4.437 ± 0.959 | 5.864 ± 0.76 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.694 ± 0.829 | 4.754 ± 1.423 |
4.12 ± 0.428 | 2.853 ± 0.946 |
7.607 ± 0.4 | 7.607 ± 1.677 |
5.864 ± 1.778 | 6.022 ± 0.182 |
2.694 ± 0.094 | 3.803 ± 0.492 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
