
Skermanella aerolata
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodospirillales; Azospirillaceae; Skermanella
Average proteome isoelectric point is 6.44
Get precalculated fractions of proteins
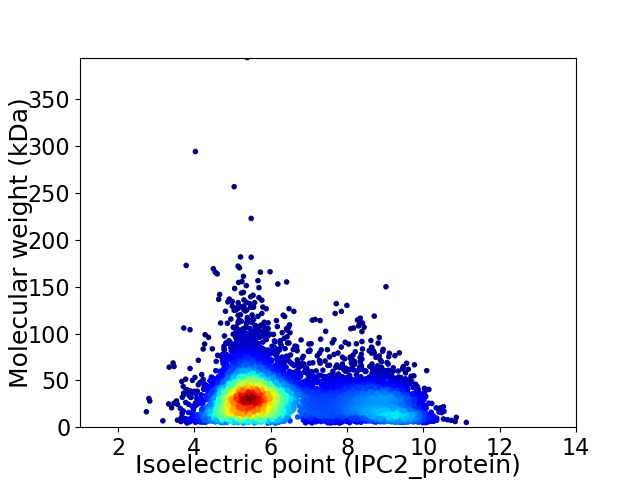
Virtual 2D-PAGE plot for 7916 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A512DNG4|A0A512DNG4_9PROT 4Fe-4S ferredoxin OS=Skermanella aerolata OX=393310 GN=SAE02_21710 PE=4 SV=1
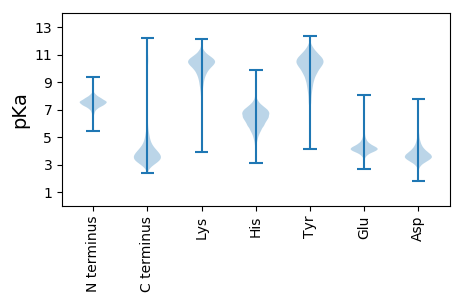
MM1 pKa = 7.83AIFNGTAARR10 pKa = 11.84DD11 pKa = 3.77IYY13 pKa = 11.15NGTSSADD20 pKa = 3.31SARR23 pKa = 11.84GNGGDD28 pKa = 4.01DD29 pKa = 3.59TLSGLGGGDD38 pKa = 3.56ALYY41 pKa = 10.96GDD43 pKa = 4.7SGNDD47 pKa = 3.42GLYY50 pKa = 10.62GGTGIDD56 pKa = 3.99RR57 pKa = 11.84IFGGTGVDD65 pKa = 3.64ALFGQDD71 pKa = 5.31GADD74 pKa = 3.37QLYY77 pKa = 10.88GEE79 pKa = 5.46AGNDD83 pKa = 3.32WLQGGAGNDD92 pKa = 3.75YY93 pKa = 11.24LHH95 pKa = 7.01GGSGNDD101 pKa = 3.7SLRR104 pKa = 11.84GDD106 pKa = 5.03DD107 pKa = 6.29GDD109 pKa = 3.98DD110 pKa = 3.64TLNGAVGDD118 pKa = 3.71NDD120 pKa = 3.59LRR122 pKa = 11.84GGAGNDD128 pKa = 3.61TLIHH132 pKa = 6.09SGIGPTYY139 pKa = 10.35SPTVNAFYY147 pKa = 10.78DD148 pKa = 3.92GGSGHH153 pKa = 7.15DD154 pKa = 4.04TLLIDD159 pKa = 3.17YY160 pKa = 8.92WGDD163 pKa = 3.63YY164 pKa = 10.04IQTSTDD170 pKa = 3.67FSPWVEE176 pKa = 3.8ININPDD182 pKa = 2.81ASSTMNYY189 pKa = 10.23SSDD192 pKa = 3.76PLEE195 pKa = 4.4GPYY198 pKa = 10.4EE199 pKa = 4.0IVAAFAGIEE208 pKa = 4.14EE209 pKa = 4.2FRR211 pKa = 11.84LTGEE215 pKa = 4.1TGRR218 pKa = 11.84LSVTARR224 pKa = 11.84TDD226 pKa = 3.26AVIFGNDD233 pKa = 3.2EE234 pKa = 3.81RR235 pKa = 11.84DD236 pKa = 3.53KK237 pKa = 11.8LEE239 pKa = 4.05GRR241 pKa = 11.84QRR243 pKa = 11.84DD244 pKa = 3.31QDD246 pKa = 3.61FTGGGGADD254 pKa = 3.15EE255 pKa = 4.41YY256 pKa = 11.03QFYY259 pKa = 8.92WRR261 pKa = 11.84KK262 pKa = 9.95GFEE265 pKa = 4.29PNHH268 pKa = 5.91DD269 pKa = 3.9TVHH272 pKa = 6.05GFSVAEE278 pKa = 4.17GDD280 pKa = 3.79TIAFGNQSEE289 pKa = 4.41NDD291 pKa = 3.58LDD293 pKa = 4.06EE294 pKa = 5.17PLPLDD299 pKa = 3.35ITAEE303 pKa = 4.21EE304 pKa = 4.73VNGHH308 pKa = 6.07TIYY311 pKa = 10.73TSVEE315 pKa = 3.64IATGQVVHH323 pKa = 6.39TLDD326 pKa = 3.44VDD328 pKa = 4.04AVGLPEE334 pKa = 4.22PQPWYY339 pKa = 10.8YY340 pKa = 10.98LGG342 pKa = 3.91
MM1 pKa = 7.83AIFNGTAARR10 pKa = 11.84DD11 pKa = 3.77IYY13 pKa = 11.15NGTSSADD20 pKa = 3.31SARR23 pKa = 11.84GNGGDD28 pKa = 4.01DD29 pKa = 3.59TLSGLGGGDD38 pKa = 3.56ALYY41 pKa = 10.96GDD43 pKa = 4.7SGNDD47 pKa = 3.42GLYY50 pKa = 10.62GGTGIDD56 pKa = 3.99RR57 pKa = 11.84IFGGTGVDD65 pKa = 3.64ALFGQDD71 pKa = 5.31GADD74 pKa = 3.37QLYY77 pKa = 10.88GEE79 pKa = 5.46AGNDD83 pKa = 3.32WLQGGAGNDD92 pKa = 3.75YY93 pKa = 11.24LHH95 pKa = 7.01GGSGNDD101 pKa = 3.7SLRR104 pKa = 11.84GDD106 pKa = 5.03DD107 pKa = 6.29GDD109 pKa = 3.98DD110 pKa = 3.64TLNGAVGDD118 pKa = 3.71NDD120 pKa = 3.59LRR122 pKa = 11.84GGAGNDD128 pKa = 3.61TLIHH132 pKa = 6.09SGIGPTYY139 pKa = 10.35SPTVNAFYY147 pKa = 10.78DD148 pKa = 3.92GGSGHH153 pKa = 7.15DD154 pKa = 4.04TLLIDD159 pKa = 3.17YY160 pKa = 8.92WGDD163 pKa = 3.63YY164 pKa = 10.04IQTSTDD170 pKa = 3.67FSPWVEE176 pKa = 3.8ININPDD182 pKa = 2.81ASSTMNYY189 pKa = 10.23SSDD192 pKa = 3.76PLEE195 pKa = 4.4GPYY198 pKa = 10.4EE199 pKa = 4.0IVAAFAGIEE208 pKa = 4.14EE209 pKa = 4.2FRR211 pKa = 11.84LTGEE215 pKa = 4.1TGRR218 pKa = 11.84LSVTARR224 pKa = 11.84TDD226 pKa = 3.26AVIFGNDD233 pKa = 3.2EE234 pKa = 3.81RR235 pKa = 11.84DD236 pKa = 3.53KK237 pKa = 11.8LEE239 pKa = 4.05GRR241 pKa = 11.84QRR243 pKa = 11.84DD244 pKa = 3.31QDD246 pKa = 3.61FTGGGGADD254 pKa = 3.15EE255 pKa = 4.41YY256 pKa = 11.03QFYY259 pKa = 8.92WRR261 pKa = 11.84KK262 pKa = 9.95GFEE265 pKa = 4.29PNHH268 pKa = 5.91DD269 pKa = 3.9TVHH272 pKa = 6.05GFSVAEE278 pKa = 4.17GDD280 pKa = 3.79TIAFGNQSEE289 pKa = 4.41NDD291 pKa = 3.58LDD293 pKa = 4.06EE294 pKa = 5.17PLPLDD299 pKa = 3.35ITAEE303 pKa = 4.21EE304 pKa = 4.73VNGHH308 pKa = 6.07TIYY311 pKa = 10.73TSVEE315 pKa = 3.64IATGQVVHH323 pKa = 6.39TLDD326 pKa = 3.44VDD328 pKa = 4.04AVGLPEE334 pKa = 4.22PQPWYY339 pKa = 10.8YY340 pKa = 10.98LGG342 pKa = 3.91
Molecular weight: 36.02 kDa
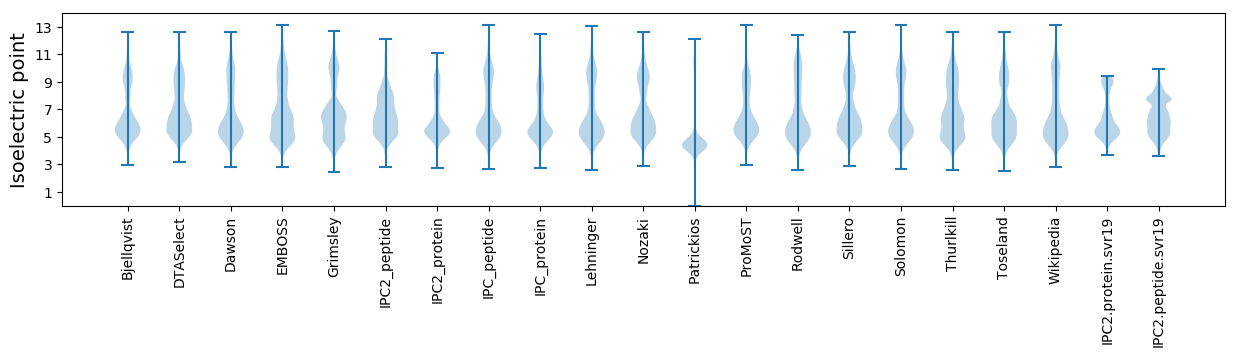
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A512DWP1|A0A512DWP1_9PROT Uncharacterized protein OS=Skermanella aerolata OX=393310 GN=SAE02_50290 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.09QPSKK9 pKa = 9.06IVRR12 pKa = 11.84KK13 pKa = 9.03HH14 pKa = 4.06RR15 pKa = 11.84HH16 pKa = 4.08GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84RR29 pKa = 11.84VLSARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.97GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.09QPSKK9 pKa = 9.06IVRR12 pKa = 11.84KK13 pKa = 9.03HH14 pKa = 4.06RR15 pKa = 11.84HH16 pKa = 4.08GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84RR29 pKa = 11.84VLSARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.97GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.13 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2418692 |
39 |
3687 |
305.5 |
33.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.977 ± 0.037 | 0.871 ± 0.009 |
5.847 ± 0.025 | 5.632 ± 0.025 |
3.545 ± 0.014 | 8.72 ± 0.025 |
2.083 ± 0.014 | 5.075 ± 0.019 |
2.986 ± 0.021 | 10.366 ± 0.034 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.531 ± 0.012 | 2.532 ± 0.015 |
5.369 ± 0.023 | 3.111 ± 0.018 |
7.35 ± 0.031 | 5.433 ± 0.017 |
5.432 ± 0.022 | 7.668 ± 0.025 |
1.324 ± 0.012 | 2.147 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
