
Trypanosoma brucei brucei (strain 927/4 GUTat10.1)
Taxonomy: cellular organisms; Eukaryota; Discoba; Euglenozoa; Kinetoplastea; Metakinetoplastina; Trypanosomatida; Trypanosomatidae; Trypanosoma; Trypanozoon; Trypanosoma brucei; Trypanosoma brucei brucei
Average proteome isoelectric point is 6.83
Get precalculated fractions of proteins
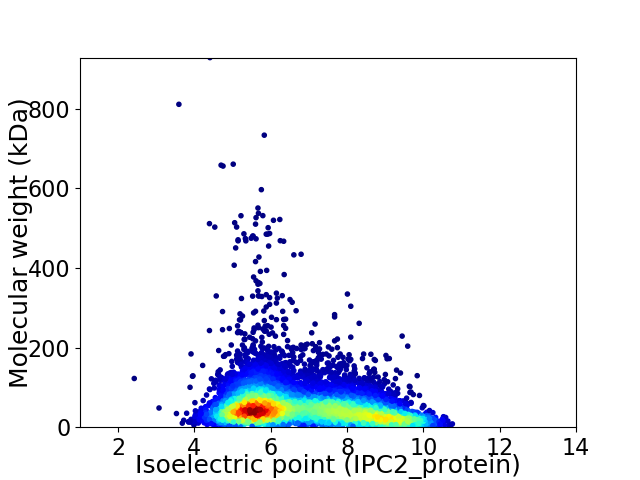
Virtual 2D-PAGE plot for 8587 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q38EE3|Q38EE3_TRYB2 Pescadillo homolog OS=Trypanosoma brucei brucei (strain 927/4 GUTat10.1) OX=185431 GN=Tb09.211.0180 PE=3 SV=1
MM1 pKa = 7.75SDD3 pKa = 4.24DD4 pKa = 4.65DD5 pKa = 5.4DD6 pKa = 5.32DD7 pKa = 4.98VVVLQVCANKK17 pKa = 9.86QCLGIEE23 pKa = 4.14DD24 pKa = 4.34LQFDD28 pKa = 3.89EE29 pKa = 6.16AMGEE33 pKa = 4.3MYY35 pKa = 10.42CANCRR40 pKa = 11.84EE41 pKa = 4.41LYY43 pKa = 10.55NRR45 pKa = 11.84AAQEE49 pKa = 3.94GFRR52 pKa = 11.84ILLSPEE58 pKa = 3.94DD59 pKa = 3.81VVLVRR64 pKa = 11.84MIFDD68 pKa = 3.71RR69 pKa = 11.84FDD71 pKa = 3.1NGKK74 pKa = 10.7GYY76 pKa = 7.36WTHH79 pKa = 7.25DD80 pKa = 3.24DD81 pKa = 3.57FVRR84 pKa = 11.84FQEE87 pKa = 4.21STLQDD92 pKa = 3.32VEE94 pKa = 4.12TDD96 pKa = 3.72INSHH100 pKa = 5.45EE101 pKa = 4.02ALKK104 pKa = 10.89EE105 pKa = 4.0FFKK108 pKa = 11.08DD109 pKa = 3.4EE110 pKa = 4.09YY111 pKa = 10.97DD112 pKa = 3.64IEE114 pKa = 4.42LTPSSSGEE122 pKa = 3.88YY123 pKa = 10.44NILPEE128 pKa = 4.52NLEE131 pKa = 4.02EE132 pKa = 4.29MYY134 pKa = 10.86GGYY137 pKa = 10.67AYY139 pKa = 11.34NNINALHH146 pKa = 6.92KK147 pKa = 11.03DD148 pKa = 3.3CDD150 pKa = 3.88ALEE153 pKa = 4.57SIGLINTAVLEE164 pKa = 4.25
MM1 pKa = 7.75SDD3 pKa = 4.24DD4 pKa = 4.65DD5 pKa = 5.4DD6 pKa = 5.32DD7 pKa = 4.98VVVLQVCANKK17 pKa = 9.86QCLGIEE23 pKa = 4.14DD24 pKa = 4.34LQFDD28 pKa = 3.89EE29 pKa = 6.16AMGEE33 pKa = 4.3MYY35 pKa = 10.42CANCRR40 pKa = 11.84EE41 pKa = 4.41LYY43 pKa = 10.55NRR45 pKa = 11.84AAQEE49 pKa = 3.94GFRR52 pKa = 11.84ILLSPEE58 pKa = 3.94DD59 pKa = 3.81VVLVRR64 pKa = 11.84MIFDD68 pKa = 3.71RR69 pKa = 11.84FDD71 pKa = 3.1NGKK74 pKa = 10.7GYY76 pKa = 7.36WTHH79 pKa = 7.25DD80 pKa = 3.24DD81 pKa = 3.57FVRR84 pKa = 11.84FQEE87 pKa = 4.21STLQDD92 pKa = 3.32VEE94 pKa = 4.12TDD96 pKa = 3.72INSHH100 pKa = 5.45EE101 pKa = 4.02ALKK104 pKa = 10.89EE105 pKa = 4.0FFKK108 pKa = 11.08DD109 pKa = 3.4EE110 pKa = 4.09YY111 pKa = 10.97DD112 pKa = 3.64IEE114 pKa = 4.42LTPSSSGEE122 pKa = 3.88YY123 pKa = 10.44NILPEE128 pKa = 4.52NLEE131 pKa = 4.02EE132 pKa = 4.29MYY134 pKa = 10.86GGYY137 pKa = 10.67AYY139 pKa = 11.34NNINALHH146 pKa = 6.92KK147 pKa = 11.03DD148 pKa = 3.3CDD150 pKa = 3.88ALEE153 pKa = 4.57SIGLINTAVLEE164 pKa = 4.25
Molecular weight: 18.81 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q584Q9|Q584Q9_TRYB2 Uncharacterized protein OS=Trypanosoma brucei brucei (strain 927/4 GUTat10.1) OX=185431 GN=Tb927.6.2470 PE=4 SV=1
MM1 pKa = 7.39NLPFPTHH8 pKa = 6.09NKK10 pKa = 8.63EE11 pKa = 3.91ALPFNEE17 pKa = 4.48DD18 pKa = 3.03AGPEE22 pKa = 4.33SRR24 pKa = 11.84SHH26 pKa = 6.79PSSDD30 pKa = 2.77SVRR33 pKa = 11.84IYY35 pKa = 10.81FSLGEE40 pKa = 3.92MAAAVSPLHH49 pKa = 6.61RR50 pKa = 11.84CFSFPVTRR58 pKa = 11.84PFVSPCIRR66 pKa = 11.84CPNCKK71 pKa = 9.81GCQLHH76 pKa = 5.82MQFCSGAYY84 pKa = 9.71GLRR87 pKa = 11.84HH88 pKa = 4.7TLAWIVRR95 pKa = 11.84RR96 pKa = 11.84HH97 pKa = 5.58PPAAEE102 pKa = 3.73EE103 pKa = 3.84RR104 pKa = 11.84RR105 pKa = 11.84TIKK108 pKa = 10.44LRR110 pKa = 11.84SQPPSAGGGAACRR123 pKa = 11.84MVRR126 pKa = 11.84RR127 pKa = 11.84VCRR130 pKa = 11.84FEE132 pKa = 3.89VAPVVITQHH141 pKa = 4.86VTHH144 pKa = 6.67VDD146 pKa = 3.33YY147 pKa = 11.48NQTIDD152 pKa = 3.44TARR155 pKa = 11.84HH156 pKa = 5.19GGSRR160 pKa = 11.84GPFAGMVCPRR170 pKa = 11.84PQEE173 pKa = 3.71IRR175 pKa = 11.84SVYY178 pKa = 10.08SRR180 pKa = 11.84IFHH183 pKa = 4.84EE184 pKa = 4.99HH185 pKa = 5.63IRR187 pKa = 11.84TGGSRR192 pKa = 11.84LPFRR196 pKa = 11.84HH197 pKa = 5.55FQRR200 pKa = 11.84TAMRR204 pKa = 11.84SLAGEE209 pKa = 4.18AQALWEE215 pKa = 4.48KK216 pKa = 10.17LTFAPEE222 pKa = 4.42GSRR225 pKa = 11.84LDD227 pKa = 3.91EE228 pKa = 4.28PPMRR232 pKa = 11.84TEE234 pKa = 4.72RR235 pKa = 11.84GDD237 pKa = 3.42KK238 pKa = 10.08VPRR241 pKa = 11.84RR242 pKa = 11.84VPIGWKK248 pKa = 9.78CPMFFVSLLPRR259 pKa = 11.84AAAGNNGITFKK270 pKa = 10.9LWLPEE275 pKa = 3.88ALVHH279 pKa = 7.4DD280 pKa = 5.39FLTAHH285 pKa = 5.96IVEE288 pKa = 4.35VEE290 pKa = 4.19RR291 pKa = 11.84VRR293 pKa = 11.84EE294 pKa = 4.02LPHH297 pKa = 5.37TCRR300 pKa = 11.84FRR302 pKa = 11.84CLSTDD307 pKa = 2.85TMLEE311 pKa = 3.93VEE313 pKa = 4.62DD314 pKa = 5.84AIASVNALLPRR325 pKa = 11.84WEE327 pKa = 5.02GEE329 pKa = 4.3LKK331 pKa = 10.66CPRR334 pKa = 11.84CYY336 pKa = 10.64GRR338 pKa = 11.84RR339 pKa = 11.84NHH341 pKa = 6.92WGFICNRR348 pKa = 11.84HH349 pKa = 5.47RR350 pKa = 11.84RR351 pKa = 11.84QLDD354 pKa = 3.67PIITLLRR361 pKa = 11.84VRR363 pKa = 11.84QSSVRR368 pKa = 11.84RR369 pKa = 11.84RR370 pKa = 11.84VQRR373 pKa = 11.84RR374 pKa = 11.84AEE376 pKa = 4.03LSGHH380 pKa = 5.17VVDD383 pKa = 5.56GAMKK387 pKa = 10.93NEE389 pKa = 4.05VVWMNKK395 pKa = 8.51RR396 pKa = 11.84KK397 pKa = 7.95TRR399 pKa = 11.84AQVMALMLANRR410 pKa = 11.84MPTTVCGPFRR420 pKa = 11.84QRR422 pKa = 11.84HH423 pKa = 5.07LPAPPSFKK431 pKa = 10.45EE432 pKa = 4.05GCARR436 pKa = 3.88
MM1 pKa = 7.39NLPFPTHH8 pKa = 6.09NKK10 pKa = 8.63EE11 pKa = 3.91ALPFNEE17 pKa = 4.48DD18 pKa = 3.03AGPEE22 pKa = 4.33SRR24 pKa = 11.84SHH26 pKa = 6.79PSSDD30 pKa = 2.77SVRR33 pKa = 11.84IYY35 pKa = 10.81FSLGEE40 pKa = 3.92MAAAVSPLHH49 pKa = 6.61RR50 pKa = 11.84CFSFPVTRR58 pKa = 11.84PFVSPCIRR66 pKa = 11.84CPNCKK71 pKa = 9.81GCQLHH76 pKa = 5.82MQFCSGAYY84 pKa = 9.71GLRR87 pKa = 11.84HH88 pKa = 4.7TLAWIVRR95 pKa = 11.84RR96 pKa = 11.84HH97 pKa = 5.58PPAAEE102 pKa = 3.73EE103 pKa = 3.84RR104 pKa = 11.84RR105 pKa = 11.84TIKK108 pKa = 10.44LRR110 pKa = 11.84SQPPSAGGGAACRR123 pKa = 11.84MVRR126 pKa = 11.84RR127 pKa = 11.84VCRR130 pKa = 11.84FEE132 pKa = 3.89VAPVVITQHH141 pKa = 4.86VTHH144 pKa = 6.67VDD146 pKa = 3.33YY147 pKa = 11.48NQTIDD152 pKa = 3.44TARR155 pKa = 11.84HH156 pKa = 5.19GGSRR160 pKa = 11.84GPFAGMVCPRR170 pKa = 11.84PQEE173 pKa = 3.71IRR175 pKa = 11.84SVYY178 pKa = 10.08SRR180 pKa = 11.84IFHH183 pKa = 4.84EE184 pKa = 4.99HH185 pKa = 5.63IRR187 pKa = 11.84TGGSRR192 pKa = 11.84LPFRR196 pKa = 11.84HH197 pKa = 5.55FQRR200 pKa = 11.84TAMRR204 pKa = 11.84SLAGEE209 pKa = 4.18AQALWEE215 pKa = 4.48KK216 pKa = 10.17LTFAPEE222 pKa = 4.42GSRR225 pKa = 11.84LDD227 pKa = 3.91EE228 pKa = 4.28PPMRR232 pKa = 11.84TEE234 pKa = 4.72RR235 pKa = 11.84GDD237 pKa = 3.42KK238 pKa = 10.08VPRR241 pKa = 11.84RR242 pKa = 11.84VPIGWKK248 pKa = 9.78CPMFFVSLLPRR259 pKa = 11.84AAAGNNGITFKK270 pKa = 10.9LWLPEE275 pKa = 3.88ALVHH279 pKa = 7.4DD280 pKa = 5.39FLTAHH285 pKa = 5.96IVEE288 pKa = 4.35VEE290 pKa = 4.19RR291 pKa = 11.84VRR293 pKa = 11.84EE294 pKa = 4.02LPHH297 pKa = 5.37TCRR300 pKa = 11.84FRR302 pKa = 11.84CLSTDD307 pKa = 2.85TMLEE311 pKa = 3.93VEE313 pKa = 4.62DD314 pKa = 5.84AIASVNALLPRR325 pKa = 11.84WEE327 pKa = 5.02GEE329 pKa = 4.3LKK331 pKa = 10.66CPRR334 pKa = 11.84CYY336 pKa = 10.64GRR338 pKa = 11.84RR339 pKa = 11.84NHH341 pKa = 6.92WGFICNRR348 pKa = 11.84HH349 pKa = 5.47RR350 pKa = 11.84RR351 pKa = 11.84QLDD354 pKa = 3.67PIITLLRR361 pKa = 11.84VRR363 pKa = 11.84QSSVRR368 pKa = 11.84RR369 pKa = 11.84RR370 pKa = 11.84VQRR373 pKa = 11.84RR374 pKa = 11.84AEE376 pKa = 4.03LSGHH380 pKa = 5.17VVDD383 pKa = 5.56GAMKK387 pKa = 10.93NEE389 pKa = 4.05VVWMNKK395 pKa = 8.51RR396 pKa = 11.84KK397 pKa = 7.95TRR399 pKa = 11.84AQVMALMLANRR410 pKa = 11.84MPTTVCGPFRR420 pKa = 11.84QRR422 pKa = 11.84HH423 pKa = 5.07LPAPPSFKK431 pKa = 10.45EE432 pKa = 4.05GCARR436 pKa = 3.88
Molecular weight: 49.62 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
4345578 |
17 |
8214 |
506.1 |
56.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.202 ± 0.029 | 2.251 ± 0.014 |
5.066 ± 0.021 | 6.994 ± 0.048 |
3.677 ± 0.017 | 6.599 ± 0.025 |
2.499 ± 0.012 | 3.998 ± 0.017 |
4.616 ± 0.025 | 9.5 ± 0.031 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.351 ± 0.011 | 3.646 ± 0.016 |
4.868 ± 0.027 | 3.833 ± 0.027 |
7.011 ± 0.022 | 8.002 ± 0.032 |
5.671 ± 0.019 | 7.585 ± 0.024 |
1.109 ± 0.009 | 2.521 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
