
Rhodococcus rhodnii LMG 5362
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Corynebacteriales; Nocardiaceae; Rhodococcus; Rhodococcus rhodnii
Average proteome isoelectric point is 6.03
Get precalculated fractions of proteins
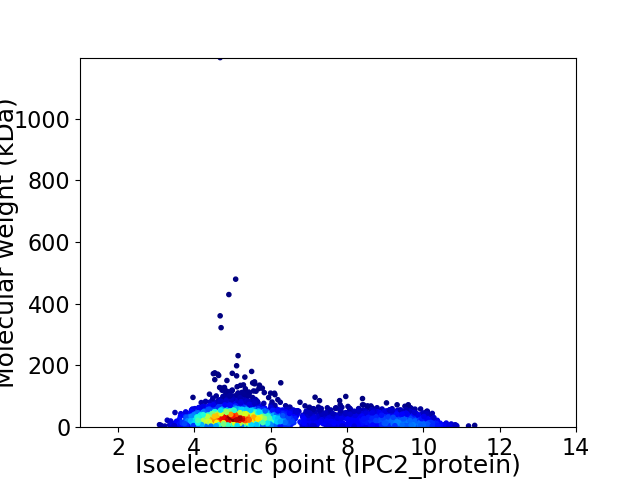
Virtual 2D-PAGE plot for 4412 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R7WI45|R7WI45_9NOCA YcaO domain-containing protein OS=Rhodococcus rhodnii LMG 5362 OX=1273125 GN=Rrhod_3826 PE=4 SV=1
MM1 pKa = 7.52ALAATTGLVLTACSGEE17 pKa = 4.27DD18 pKa = 3.14AATASSGYY26 pKa = 9.81DD27 pKa = 3.55GPIGAIDD34 pKa = 4.77LSQDD38 pKa = 3.27CPATVVIQTDD48 pKa = 3.38WNPEE52 pKa = 3.89VEE54 pKa = 4.69HH55 pKa = 6.35GGSYY59 pKa = 10.88QMLGPDD65 pKa = 3.74PQIDD69 pKa = 3.86ANSKK73 pKa = 8.36RR74 pKa = 11.84TSGPLFASGEE84 pKa = 4.22YY85 pKa = 9.85TGVDD89 pKa = 3.31LEE91 pKa = 4.39IRR93 pKa = 11.84SGGPAIGFQSVTSQMYY109 pKa = 9.35QDD111 pKa = 4.1PDD113 pKa = 3.08IMLGYY118 pKa = 10.37VDD120 pKa = 4.09TDD122 pKa = 3.49QAISNSGNNPTIGVYY137 pKa = 9.93AALEE141 pKa = 4.17KK142 pKa = 10.96SPMMLMWDD150 pKa = 3.7PEE152 pKa = 4.41TYY154 pKa = 10.31PDD156 pKa = 3.57VEE158 pKa = 4.67RR159 pKa = 11.84IEE161 pKa = 4.63DD162 pKa = 4.41LKK164 pKa = 10.92DD165 pKa = 3.33TPAKK169 pKa = 9.91VLYY172 pKa = 10.33FEE174 pKa = 5.42GEE176 pKa = 3.75AYY178 pKa = 9.52MDD180 pKa = 4.09YY181 pKa = 10.23LTGSGVLNPDD191 pKa = 3.47QVDD194 pKa = 3.61GSYY197 pKa = 11.23DD198 pKa = 3.36GTPANFVAAGGANGQQGFASSEE220 pKa = 4.05PYY222 pKa = 10.01VYY224 pKa = 10.6EE225 pKa = 4.48NEE227 pKa = 4.08VEE229 pKa = 4.37GWKK232 pKa = 10.62KK233 pKa = 9.83PVNFEE238 pKa = 4.92LIHH241 pKa = 5.89DD242 pKa = 4.36TGYY245 pKa = 8.57PTYY248 pKa = 10.33KK249 pKa = 10.45SPLAVRR255 pKa = 11.84ADD257 pKa = 3.46ALEE260 pKa = 4.39EE261 pKa = 3.88NSACLEE267 pKa = 3.95KK268 pKa = 10.59LVPVMQQASVDD279 pKa = 3.96YY280 pKa = 10.23FADD283 pKa = 3.86PAAANAVILDD293 pKa = 3.58AVEE296 pKa = 4.04QYY298 pKa = 8.91NTGWVYY304 pKa = 10.79GQGNADD310 pKa = 3.7YY311 pKa = 10.51GLEE314 pKa = 4.05TMTEE318 pKa = 4.13LEE320 pKa = 4.18LVGNGTTPTHH330 pKa = 6.51GDD332 pKa = 3.01FDD334 pKa = 4.15EE335 pKa = 5.78ARR337 pKa = 11.84VQQLIDD343 pKa = 2.94ITRR346 pKa = 11.84PVFNEE351 pKa = 3.63QGKK354 pKa = 9.66DD355 pKa = 3.6VPADD359 pKa = 3.9LAPDD363 pKa = 3.4AVVTNRR369 pKa = 11.84FIDD372 pKa = 3.8EE373 pKa = 4.75SISFPP378 pKa = 4.22
MM1 pKa = 7.52ALAATTGLVLTACSGEE17 pKa = 4.27DD18 pKa = 3.14AATASSGYY26 pKa = 9.81DD27 pKa = 3.55GPIGAIDD34 pKa = 4.77LSQDD38 pKa = 3.27CPATVVIQTDD48 pKa = 3.38WNPEE52 pKa = 3.89VEE54 pKa = 4.69HH55 pKa = 6.35GGSYY59 pKa = 10.88QMLGPDD65 pKa = 3.74PQIDD69 pKa = 3.86ANSKK73 pKa = 8.36RR74 pKa = 11.84TSGPLFASGEE84 pKa = 4.22YY85 pKa = 9.85TGVDD89 pKa = 3.31LEE91 pKa = 4.39IRR93 pKa = 11.84SGGPAIGFQSVTSQMYY109 pKa = 9.35QDD111 pKa = 4.1PDD113 pKa = 3.08IMLGYY118 pKa = 10.37VDD120 pKa = 4.09TDD122 pKa = 3.49QAISNSGNNPTIGVYY137 pKa = 9.93AALEE141 pKa = 4.17KK142 pKa = 10.96SPMMLMWDD150 pKa = 3.7PEE152 pKa = 4.41TYY154 pKa = 10.31PDD156 pKa = 3.57VEE158 pKa = 4.67RR159 pKa = 11.84IEE161 pKa = 4.63DD162 pKa = 4.41LKK164 pKa = 10.92DD165 pKa = 3.33TPAKK169 pKa = 9.91VLYY172 pKa = 10.33FEE174 pKa = 5.42GEE176 pKa = 3.75AYY178 pKa = 9.52MDD180 pKa = 4.09YY181 pKa = 10.23LTGSGVLNPDD191 pKa = 3.47QVDD194 pKa = 3.61GSYY197 pKa = 11.23DD198 pKa = 3.36GTPANFVAAGGANGQQGFASSEE220 pKa = 4.05PYY222 pKa = 10.01VYY224 pKa = 10.6EE225 pKa = 4.48NEE227 pKa = 4.08VEE229 pKa = 4.37GWKK232 pKa = 10.62KK233 pKa = 9.83PVNFEE238 pKa = 4.92LIHH241 pKa = 5.89DD242 pKa = 4.36TGYY245 pKa = 8.57PTYY248 pKa = 10.33KK249 pKa = 10.45SPLAVRR255 pKa = 11.84ADD257 pKa = 3.46ALEE260 pKa = 4.39EE261 pKa = 3.88NSACLEE267 pKa = 3.95KK268 pKa = 10.59LVPVMQQASVDD279 pKa = 3.96YY280 pKa = 10.23FADD283 pKa = 3.86PAAANAVILDD293 pKa = 3.58AVEE296 pKa = 4.04QYY298 pKa = 8.91NTGWVYY304 pKa = 10.79GQGNADD310 pKa = 3.7YY311 pKa = 10.51GLEE314 pKa = 4.05TMTEE318 pKa = 4.13LEE320 pKa = 4.18LVGNGTTPTHH330 pKa = 6.51GDD332 pKa = 3.01FDD334 pKa = 4.15EE335 pKa = 5.78ARR337 pKa = 11.84VQQLIDD343 pKa = 2.94ITRR346 pKa = 11.84PVFNEE351 pKa = 3.63QGKK354 pKa = 9.66DD355 pKa = 3.6VPADD359 pKa = 3.9LAPDD363 pKa = 3.4AVVTNRR369 pKa = 11.84FIDD372 pKa = 3.8EE373 pKa = 4.75SISFPP378 pKa = 4.22
Molecular weight: 40.49 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R7WK57|R7WK57_9NOCA Uncharacterized protein OS=Rhodococcus rhodnii LMG 5362 OX=1273125 GN=Rrhod_2940 PE=4 SV=1
MM1 pKa = 6.73FTVAKK6 pKa = 9.94GKK8 pKa = 8.69RR9 pKa = 11.84TFQPNNRR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84ARR20 pKa = 11.84VHH22 pKa = 5.99GFRR25 pKa = 11.84LRR27 pKa = 11.84MRR29 pKa = 11.84TRR31 pKa = 11.84AGRR34 pKa = 11.84AIVTARR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84KK43 pKa = 9.64GRR45 pKa = 11.84ASLTAA50 pKa = 4.1
MM1 pKa = 6.73FTVAKK6 pKa = 9.94GKK8 pKa = 8.69RR9 pKa = 11.84TFQPNNRR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84ARR20 pKa = 11.84VHH22 pKa = 5.99GFRR25 pKa = 11.84LRR27 pKa = 11.84MRR29 pKa = 11.84TRR31 pKa = 11.84AGRR34 pKa = 11.84AIVTARR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84KK43 pKa = 9.64GRR45 pKa = 11.84ASLTAA50 pKa = 4.1
Molecular weight: 5.84 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1323253 |
30 |
11261 |
299.9 |
32.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.45 ± 0.053 | 0.713 ± 0.01 |
6.725 ± 0.035 | 5.57 ± 0.031 |
2.916 ± 0.022 | 9.175 ± 0.033 |
2.169 ± 0.019 | 3.832 ± 0.025 |
1.676 ± 0.029 | 9.608 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.866 ± 0.015 | 1.735 ± 0.018 |
5.724 ± 0.032 | 2.434 ± 0.021 |
7.899 ± 0.044 | 5.499 ± 0.027 |
6.31 ± 0.027 | 9.34 ± 0.041 |
1.437 ± 0.016 | 1.923 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
