
Bradyrhizobium diazoefficiens (strain JCM 10833 / BCRC 13528 / IAM 13628 / NBRC 14792 / USDA 110)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Bradyrhizobiaceae; Bradyrhizobium; Bradyrhizobium diazoefficiens
Average proteome isoelectric point is 7.07
Get precalculated fractions of proteins
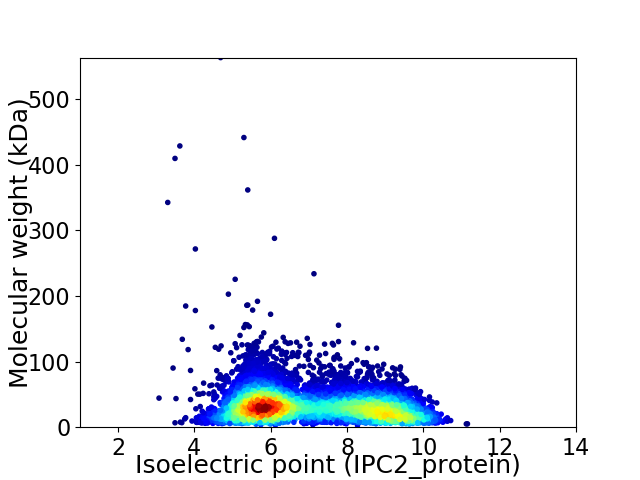
Virtual 2D-PAGE plot for 8253 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q89DD5|Q89DD5_BRADU Bsl7504 protein OS=Bradyrhizobium diazoefficiens (strain JCM 10833 / BCRC 13528 / IAM 13628 / NBRC 14792 / USDA 110) OX=224911 GN=bsl7504 PE=4 SV=1
MM1 pKa = 7.13TVIPPASGMTLHH13 pKa = 7.4DD14 pKa = 5.02GDD16 pKa = 4.31TMDD19 pKa = 3.14VSGGISVDD27 pKa = 3.1STTILAGALQTVGGSFATSTATHH50 pKa = 5.34TVINGGEE57 pKa = 3.89QDD59 pKa = 3.86VVFDD63 pKa = 4.34GVASYY68 pKa = 7.43TTINAGGVQRR78 pKa = 11.84VTGEE82 pKa = 4.24YY83 pKa = 9.0IHH85 pKa = 6.75EE86 pKa = 4.79GPTPGEE92 pKa = 4.01VDD94 pKa = 2.97HH95 pKa = 6.07TTINGGGEE103 pKa = 3.94QDD105 pKa = 3.58VDD107 pKa = 3.47TGIATTTTVNSGGVQNVTNGGSASGTIVNSGGVLRR142 pKa = 11.84VSGEE146 pKa = 4.32TVEE149 pKa = 4.31TAPVYY154 pKa = 9.49PEE156 pKa = 3.57PVIRR160 pKa = 11.84SVATAATINYY170 pKa = 9.27QGEE173 pKa = 4.41LDD175 pKa = 3.77VTGGGTAVNAIVNGGVMTVSGSIPDD200 pKa = 3.87PFANVPGQPAVDD212 pKa = 3.35ASMATGTIVEE222 pKa = 4.55GFGTLTVSDD231 pKa = 4.22GALVSGTILQNFGTLNVDD249 pKa = 3.41AGGTATGTTINYY261 pKa = 7.19GTMVLSAGATAINTTVNQYY280 pKa = 10.75GGLVVLGNADD290 pKa = 3.38FSGTVFNAGSTLQIGDD306 pKa = 3.62GTIEE310 pKa = 4.19NGFTVPKK317 pKa = 10.24SVTLAVLDD325 pKa = 4.19GGVLSNSVISAGATLIAEE343 pKa = 4.92AGATLSNTTFQSGATFVADD362 pKa = 3.43YY363 pKa = 10.25GYY365 pKa = 9.08TEE367 pKa = 4.15NGFTVSDD374 pKa = 4.77GILFKK379 pKa = 11.43VFGTATNTTISAGGKK394 pKa = 9.61QILGNYY400 pKa = 9.32DD401 pKa = 3.63LGATASNTVINGGEE415 pKa = 3.91QDD417 pKa = 3.56VGLASTASYY426 pKa = 7.77TTINTGGIQRR436 pKa = 11.84VAGIYY441 pKa = 10.29LHH443 pKa = 7.18DD444 pKa = 4.5APGPGRR450 pKa = 11.84ADD452 pKa = 2.85HH453 pKa = 5.78TTVNNGAEE461 pKa = 4.13QDD463 pKa = 3.42VDD465 pKa = 3.67TGIVTSTTVNGGGVQNLTNGGFASGTVVNSGGVINASGEE504 pKa = 4.26TIYY507 pKa = 10.81YY508 pKa = 9.23PPGGPYY514 pKa = 10.46GIVFRR519 pKa = 11.84SEE521 pKa = 3.07VDD523 pKa = 3.9FAVVGSGGQLHH534 pKa = 6.89LFGAGIAKK542 pKa = 9.87KK543 pKa = 10.41AIINGGVMTVSGSVADD559 pKa = 3.96PTNDD563 pKa = 3.22PSAPAVDD570 pKa = 4.57ASAASGTVLNSGSLSVSDD588 pKa = 3.95GGIVTSTILNGGTLDD603 pKa = 4.4VLDD606 pKa = 4.28QGTANASTVLRR617 pKa = 11.84GATEE621 pKa = 3.88LVEE624 pKa = 4.55AGGIAGATTIAGGMLDD640 pKa = 4.62LKK642 pKa = 11.01AGAISDD648 pKa = 3.95DD649 pKa = 4.6AITFSGQGTLKK660 pKa = 10.2IEE662 pKa = 4.14QKK664 pKa = 10.54LVSGSSAFASQIKK677 pKa = 10.27GIALGDD683 pKa = 3.84QIDD686 pKa = 4.18LSGLSYY692 pKa = 10.88TSGATVTVSGSKK704 pKa = 9.15LTVSNGAASEE714 pKa = 4.34TFTLADD720 pKa = 3.33TSVTRR725 pKa = 11.84FGIAKK730 pKa = 9.66DD731 pKa = 3.51SSGGILLTAAALPAIAGAVSGQATSNQASINPFATVTITDD771 pKa = 4.0PNAAATDD778 pKa = 3.78SLIITLAGAAGILSGSGLISLGNGTYY804 pKa = 10.56EE805 pKa = 4.61LAATSAANLTAEE817 pKa = 4.48LHH819 pKa = 6.31ALTFVASPHH828 pKa = 6.4GDD830 pKa = 3.0GSVTTTFTLSDD841 pKa = 3.6TSSVGLTVSDD851 pKa = 3.95TNTSVTDD858 pKa = 2.99THH860 pKa = 6.44QAGTTTIDD868 pKa = 3.73GPASGYY874 pKa = 11.09AIIQGTTGNDD884 pKa = 3.04IIHH887 pKa = 7.26AYY889 pKa = 10.79GMFNTIHH896 pKa = 6.69GNGGSDD902 pKa = 3.56TIYY905 pKa = 10.54AGLYY909 pKa = 7.63GTVDD913 pKa = 3.62VPSGDD918 pKa = 3.52SAVYY922 pKa = 9.65LQGIVNHH929 pKa = 5.25VTGGYY934 pKa = 10.89GNVTVTGSTGATTIALGNGNDD955 pKa = 4.03TIDD958 pKa = 3.2ASGYY962 pKa = 10.58GNVITLGNGNDD973 pKa = 3.4IVHH976 pKa = 7.1PGDD979 pKa = 3.89GASQTTAGNGNDD991 pKa = 3.63LVTLSGYY998 pKa = 11.1GNTVMLGNGNDD1009 pKa = 3.42VVAGRR1014 pKa = 11.84DD1015 pKa = 3.45GANSVTLGDD1024 pKa = 4.03GNNTVNLGGMGNQITVGSGTNAIIAGSGSDD1054 pKa = 3.46NVVAGAGHH1062 pKa = 6.01DD1063 pKa = 3.82TIMLGGAANHH1073 pKa = 5.65VVLNGSQANVTNQIGQDD1090 pKa = 3.38VVTVNGGSDD1099 pKa = 3.06QFNFVGFGNQAIINGPAHH1117 pKa = 6.79VDD1119 pKa = 3.67IIDD1122 pKa = 3.94HH1123 pKa = 5.7GTGLTVSVNSSSQVDD1138 pKa = 4.18TISGFGNDD1146 pKa = 3.33TLGIVALANGVGDD1159 pKa = 3.86YY1160 pKa = 11.06HH1161 pKa = 8.24SVADD1165 pKa = 3.73IMSALRR1171 pKa = 11.84SDD1173 pKa = 3.74GHH1175 pKa = 6.51GGTLLALGSGSNAGSIDD1192 pKa = 3.72FVDD1195 pKa = 3.73TAISQLHH1202 pKa = 6.58ASNFMIVV1209 pKa = 2.81
MM1 pKa = 7.13TVIPPASGMTLHH13 pKa = 7.4DD14 pKa = 5.02GDD16 pKa = 4.31TMDD19 pKa = 3.14VSGGISVDD27 pKa = 3.1STTILAGALQTVGGSFATSTATHH50 pKa = 5.34TVINGGEE57 pKa = 3.89QDD59 pKa = 3.86VVFDD63 pKa = 4.34GVASYY68 pKa = 7.43TTINAGGVQRR78 pKa = 11.84VTGEE82 pKa = 4.24YY83 pKa = 9.0IHH85 pKa = 6.75EE86 pKa = 4.79GPTPGEE92 pKa = 4.01VDD94 pKa = 2.97HH95 pKa = 6.07TTINGGGEE103 pKa = 3.94QDD105 pKa = 3.58VDD107 pKa = 3.47TGIATTTTVNSGGVQNVTNGGSASGTIVNSGGVLRR142 pKa = 11.84VSGEE146 pKa = 4.32TVEE149 pKa = 4.31TAPVYY154 pKa = 9.49PEE156 pKa = 3.57PVIRR160 pKa = 11.84SVATAATINYY170 pKa = 9.27QGEE173 pKa = 4.41LDD175 pKa = 3.77VTGGGTAVNAIVNGGVMTVSGSIPDD200 pKa = 3.87PFANVPGQPAVDD212 pKa = 3.35ASMATGTIVEE222 pKa = 4.55GFGTLTVSDD231 pKa = 4.22GALVSGTILQNFGTLNVDD249 pKa = 3.41AGGTATGTTINYY261 pKa = 7.19GTMVLSAGATAINTTVNQYY280 pKa = 10.75GGLVVLGNADD290 pKa = 3.38FSGTVFNAGSTLQIGDD306 pKa = 3.62GTIEE310 pKa = 4.19NGFTVPKK317 pKa = 10.24SVTLAVLDD325 pKa = 4.19GGVLSNSVISAGATLIAEE343 pKa = 4.92AGATLSNTTFQSGATFVADD362 pKa = 3.43YY363 pKa = 10.25GYY365 pKa = 9.08TEE367 pKa = 4.15NGFTVSDD374 pKa = 4.77GILFKK379 pKa = 11.43VFGTATNTTISAGGKK394 pKa = 9.61QILGNYY400 pKa = 9.32DD401 pKa = 3.63LGATASNTVINGGEE415 pKa = 3.91QDD417 pKa = 3.56VGLASTASYY426 pKa = 7.77TTINTGGIQRR436 pKa = 11.84VAGIYY441 pKa = 10.29LHH443 pKa = 7.18DD444 pKa = 4.5APGPGRR450 pKa = 11.84ADD452 pKa = 2.85HH453 pKa = 5.78TTVNNGAEE461 pKa = 4.13QDD463 pKa = 3.42VDD465 pKa = 3.67TGIVTSTTVNGGGVQNLTNGGFASGTVVNSGGVINASGEE504 pKa = 4.26TIYY507 pKa = 10.81YY508 pKa = 9.23PPGGPYY514 pKa = 10.46GIVFRR519 pKa = 11.84SEE521 pKa = 3.07VDD523 pKa = 3.9FAVVGSGGQLHH534 pKa = 6.89LFGAGIAKK542 pKa = 9.87KK543 pKa = 10.41AIINGGVMTVSGSVADD559 pKa = 3.96PTNDD563 pKa = 3.22PSAPAVDD570 pKa = 4.57ASAASGTVLNSGSLSVSDD588 pKa = 3.95GGIVTSTILNGGTLDD603 pKa = 4.4VLDD606 pKa = 4.28QGTANASTVLRR617 pKa = 11.84GATEE621 pKa = 3.88LVEE624 pKa = 4.55AGGIAGATTIAGGMLDD640 pKa = 4.62LKK642 pKa = 11.01AGAISDD648 pKa = 3.95DD649 pKa = 4.6AITFSGQGTLKK660 pKa = 10.2IEE662 pKa = 4.14QKK664 pKa = 10.54LVSGSSAFASQIKK677 pKa = 10.27GIALGDD683 pKa = 3.84QIDD686 pKa = 4.18LSGLSYY692 pKa = 10.88TSGATVTVSGSKK704 pKa = 9.15LTVSNGAASEE714 pKa = 4.34TFTLADD720 pKa = 3.33TSVTRR725 pKa = 11.84FGIAKK730 pKa = 9.66DD731 pKa = 3.51SSGGILLTAAALPAIAGAVSGQATSNQASINPFATVTITDD771 pKa = 4.0PNAAATDD778 pKa = 3.78SLIITLAGAAGILSGSGLISLGNGTYY804 pKa = 10.56EE805 pKa = 4.61LAATSAANLTAEE817 pKa = 4.48LHH819 pKa = 6.31ALTFVASPHH828 pKa = 6.4GDD830 pKa = 3.0GSVTTTFTLSDD841 pKa = 3.6TSSVGLTVSDD851 pKa = 3.95TNTSVTDD858 pKa = 2.99THH860 pKa = 6.44QAGTTTIDD868 pKa = 3.73GPASGYY874 pKa = 11.09AIIQGTTGNDD884 pKa = 3.04IIHH887 pKa = 7.26AYY889 pKa = 10.79GMFNTIHH896 pKa = 6.69GNGGSDD902 pKa = 3.56TIYY905 pKa = 10.54AGLYY909 pKa = 7.63GTVDD913 pKa = 3.62VPSGDD918 pKa = 3.52SAVYY922 pKa = 9.65LQGIVNHH929 pKa = 5.25VTGGYY934 pKa = 10.89GNVTVTGSTGATTIALGNGNDD955 pKa = 4.03TIDD958 pKa = 3.2ASGYY962 pKa = 10.58GNVITLGNGNDD973 pKa = 3.4IVHH976 pKa = 7.1PGDD979 pKa = 3.89GASQTTAGNGNDD991 pKa = 3.63LVTLSGYY998 pKa = 11.1GNTVMLGNGNDD1009 pKa = 3.42VVAGRR1014 pKa = 11.84DD1015 pKa = 3.45GANSVTLGDD1024 pKa = 4.03GNNTVNLGGMGNQITVGSGTNAIIAGSGSDD1054 pKa = 3.46NVVAGAGHH1062 pKa = 6.01DD1063 pKa = 3.82TIMLGGAANHH1073 pKa = 5.65VVLNGSQANVTNQIGQDD1090 pKa = 3.38VVTVNGGSDD1099 pKa = 3.06QFNFVGFGNQAIINGPAHH1117 pKa = 6.79VDD1119 pKa = 3.67IIDD1122 pKa = 3.94HH1123 pKa = 5.7GTGLTVSVNSSSQVDD1138 pKa = 4.18TISGFGNDD1146 pKa = 3.33TLGIVALANGVGDD1159 pKa = 3.86YY1160 pKa = 11.06HH1161 pKa = 8.24SVADD1165 pKa = 3.73IMSALRR1171 pKa = 11.84SDD1173 pKa = 3.74GHH1175 pKa = 6.51GGTLLALGSGSNAGSIDD1192 pKa = 3.72FVDD1195 pKa = 3.73TAISQLHH1202 pKa = 6.58ASNFMIVV1209 pKa = 2.81
Molecular weight: 118.29 kDa
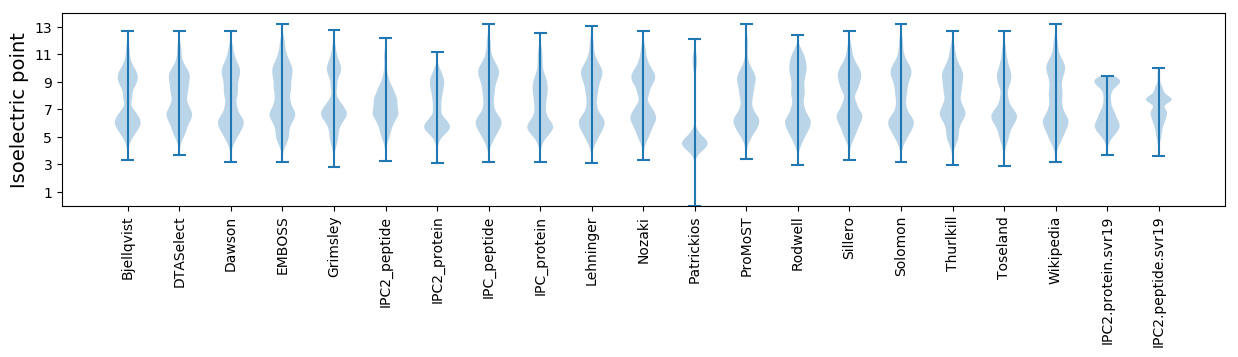
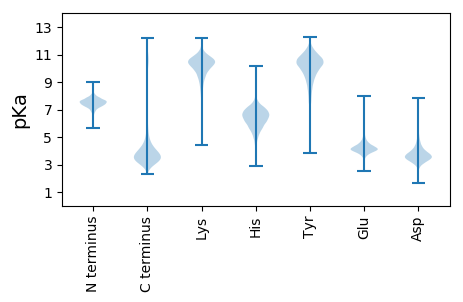
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q89BY5|Q89BY5_BRADU N-(5'-phosphoribosyl)anthranilate isomerase OS=Bradyrhizobium diazoefficiens (strain JCM 10833 / BCRC 13528 / IAM 13628 / NBRC 14792 / USDA 110) OX=224911 GN=trpF PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.55GFRR19 pKa = 11.84ARR21 pKa = 11.84LATAGGRR28 pKa = 11.84KK29 pKa = 8.95VLAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.55GFRR19 pKa = 11.84ARR21 pKa = 11.84LATAGGRR28 pKa = 11.84KK29 pKa = 8.95VLAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
Molecular weight: 5.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2616478 |
26 |
5685 |
317.0 |
34.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.422 ± 0.033 | 0.919 ± 0.01 |
5.466 ± 0.025 | 5.257 ± 0.031 |
3.737 ± 0.018 | 8.359 ± 0.031 |
2.057 ± 0.012 | 5.266 ± 0.022 |
3.609 ± 0.027 | 9.86 ± 0.035 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.467 ± 0.014 | 2.76 ± 0.022 |
5.263 ± 0.026 | 3.187 ± 0.017 |
7.186 ± 0.039 | 5.819 ± 0.024 |
5.425 ± 0.038 | 7.416 ± 0.023 |
1.316 ± 0.012 | 2.209 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
