
Ferrimonas sediminum
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Ferrimonadaceae; Ferrimonas
Average proteome isoelectric point is 6.23
Get precalculated fractions of proteins
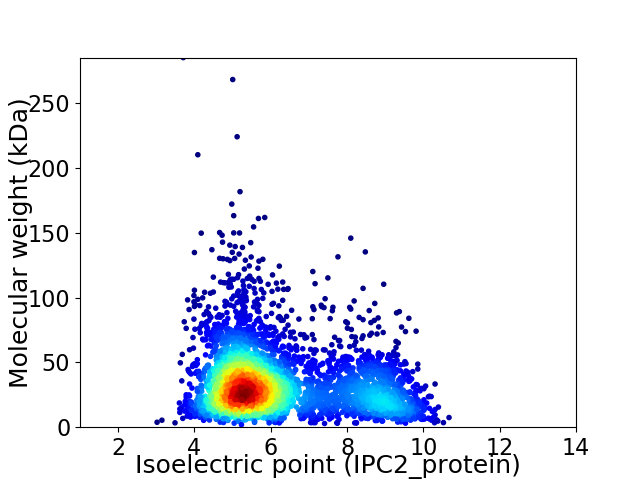
Virtual 2D-PAGE plot for 3939 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1G8Q5R6|A0A1G8Q5R6_9GAMM Positive regulator of sigma(E) RseC/MucC OS=Ferrimonas sediminum OX=718193 GN=SAMN04488540_104191 PE=4 SV=1
MM1 pKa = 7.38KK2 pKa = 10.12RR3 pKa = 11.84HH4 pKa = 5.46KK5 pKa = 9.35TNAITRR11 pKa = 11.84TLGIALVGTAGLYY24 pKa = 10.63CLVSPTTARR33 pKa = 11.84ADD35 pKa = 3.81FFPQQLNQACMAEE48 pKa = 3.81AAGFNLNCTANDD60 pKa = 3.4IQVSQVTNITRR71 pKa = 11.84PDD73 pKa = 3.62GSPGPVEE80 pKa = 4.07CTLGGDD86 pKa = 3.61AEE88 pKa = 4.33FRR90 pKa = 11.84ADD92 pKa = 3.45VTIVTTAKK100 pKa = 9.84RR101 pKa = 11.84RR102 pKa = 11.84YY103 pKa = 9.63DD104 pKa = 3.43YY105 pKa = 10.79TVYY108 pKa = 10.54LPEE111 pKa = 5.14GNWSPQEE118 pKa = 3.98FNTSNTCSVLVGQDD132 pKa = 3.03FTAPGEE138 pKa = 4.13DD139 pKa = 4.33LEE141 pKa = 5.99DD142 pKa = 4.72PGDD145 pKa = 3.64MCADD149 pKa = 3.2ISKK152 pKa = 10.26PPGSHH157 pKa = 7.53LYY159 pKa = 7.88TQQLITLNCVDD170 pKa = 4.25EE171 pKa = 5.03DD172 pKa = 3.74QDD174 pKa = 3.31GRR176 pKa = 11.84AEE178 pKa = 4.0FRR180 pKa = 11.84YY181 pKa = 9.64CAAWDD186 pKa = 3.74NKK188 pKa = 10.68ADD190 pKa = 3.85STPVCDD196 pKa = 3.85GTAATTPVPGTPSKK210 pKa = 10.7CRR212 pKa = 11.84CDD214 pKa = 3.58EE215 pKa = 4.04FNIDD219 pKa = 4.19VFIKK223 pKa = 10.28PDD225 pKa = 3.62PPTINKK231 pKa = 9.74SEE233 pKa = 4.11GSPITRR239 pKa = 11.84PEE241 pKa = 3.88PGGDD245 pKa = 3.46YY246 pKa = 10.71TFTASFTNNSLTSIFIDD263 pKa = 3.74SVADD267 pKa = 4.44EE268 pKa = 4.53IDD270 pKa = 3.38VGGEE274 pKa = 3.92GSFDD278 pKa = 3.55VSLDD282 pKa = 3.69LLNGATQTVDD292 pKa = 3.26PTNTATAEE300 pKa = 4.12GVYY303 pKa = 10.23LKK305 pKa = 10.72SLSASCAQPADD316 pKa = 3.42IGNGAGEE323 pKa = 4.44LVSGATYY330 pKa = 10.63SCTFTVTIVDD340 pKa = 3.46RR341 pKa = 11.84DD342 pKa = 3.82LPNDD346 pKa = 3.63QSPEE350 pKa = 3.99LYY352 pKa = 10.42KK353 pKa = 10.95DD354 pKa = 3.68VIKK357 pKa = 11.06VALTDD362 pKa = 3.6KK363 pKa = 11.05NGNPVVNGATCPADD377 pKa = 3.92LSPIAGDD384 pKa = 3.29HH385 pKa = 6.26CSVLRR390 pKa = 11.84TVDD393 pKa = 3.52VTNLPPSITVSKK405 pKa = 9.51TPSPDD410 pKa = 3.06QVLEE414 pKa = 4.09PGGDD418 pKa = 3.33VTFDD422 pKa = 3.0IVVTSTSGNYY432 pKa = 9.69DD433 pKa = 3.51DD434 pKa = 5.89PLTLTSLMDD443 pKa = 3.67TVFGDD448 pKa = 3.75LNGVGTCATGGSFSLAAPYY467 pKa = 10.35SCSFTEE473 pKa = 4.78TISGNAGDD481 pKa = 3.79VHH483 pKa = 7.0NNTVTAKK490 pKa = 9.97ATDD493 pKa = 3.69NEE495 pKa = 3.93NDD497 pKa = 3.23EE498 pKa = 4.66AQNSDD503 pKa = 3.16GATVNINDD511 pKa = 3.72VPSIITLVKK520 pKa = 8.67TADD523 pKa = 4.43PIEE526 pKa = 4.16VDD528 pKa = 3.57EE529 pKa = 5.05TGDD532 pKa = 3.64DD533 pKa = 3.47PTVFRR538 pKa = 11.84DD539 pKa = 3.0VDD541 pKa = 4.04FTFEE545 pKa = 4.08FSVDD549 pKa = 3.1AAGVDD554 pKa = 3.55DD555 pKa = 3.95VTFNSLTDD563 pKa = 3.95TIFGTLTGEE572 pKa = 4.41CLVDD576 pKa = 3.6TKK578 pKa = 10.95NGSPIGSTPLSGFVLQPGEE597 pKa = 4.45DD598 pKa = 3.61ASCTITKK605 pKa = 10.14ALQGDD610 pKa = 3.82AGDD613 pKa = 3.61VHH615 pKa = 7.69NNVATIKK622 pKa = 11.11GIDD625 pKa = 3.82EE626 pKa = 4.52DD627 pKa = 4.32AQPVMDD633 pKa = 5.29SDD635 pKa = 4.19DD636 pKa = 3.68ATVTFLDD643 pKa = 3.71TGLDD647 pKa = 3.63MEE649 pKa = 4.74QEE651 pKa = 4.44FAAKK655 pKa = 9.45MIAFVRR661 pKa = 11.84ITNGNVDD668 pKa = 3.68TASITGVTIKK678 pKa = 10.61GVNLVAGGGIPGQFEE693 pKa = 4.33VQNDD697 pKa = 3.92AGTSSYY703 pKa = 11.54NPGDD707 pKa = 3.56GPYY710 pKa = 9.83AFCSTGVDD718 pKa = 3.31ILPGDD723 pKa = 3.83TYY725 pKa = 11.69EE726 pKa = 4.9CAFTLKK732 pKa = 10.12LFPGFPPGDD741 pKa = 3.52INFSATGANGLVFTYY756 pKa = 10.51QDD758 pKa = 3.38NEE760 pKa = 4.48GNSVTTNVDD769 pKa = 3.5MNVMTIEE776 pKa = 3.87
MM1 pKa = 7.38KK2 pKa = 10.12RR3 pKa = 11.84HH4 pKa = 5.46KK5 pKa = 9.35TNAITRR11 pKa = 11.84TLGIALVGTAGLYY24 pKa = 10.63CLVSPTTARR33 pKa = 11.84ADD35 pKa = 3.81FFPQQLNQACMAEE48 pKa = 3.81AAGFNLNCTANDD60 pKa = 3.4IQVSQVTNITRR71 pKa = 11.84PDD73 pKa = 3.62GSPGPVEE80 pKa = 4.07CTLGGDD86 pKa = 3.61AEE88 pKa = 4.33FRR90 pKa = 11.84ADD92 pKa = 3.45VTIVTTAKK100 pKa = 9.84RR101 pKa = 11.84RR102 pKa = 11.84YY103 pKa = 9.63DD104 pKa = 3.43YY105 pKa = 10.79TVYY108 pKa = 10.54LPEE111 pKa = 5.14GNWSPQEE118 pKa = 3.98FNTSNTCSVLVGQDD132 pKa = 3.03FTAPGEE138 pKa = 4.13DD139 pKa = 4.33LEE141 pKa = 5.99DD142 pKa = 4.72PGDD145 pKa = 3.64MCADD149 pKa = 3.2ISKK152 pKa = 10.26PPGSHH157 pKa = 7.53LYY159 pKa = 7.88TQQLITLNCVDD170 pKa = 4.25EE171 pKa = 5.03DD172 pKa = 3.74QDD174 pKa = 3.31GRR176 pKa = 11.84AEE178 pKa = 4.0FRR180 pKa = 11.84YY181 pKa = 9.64CAAWDD186 pKa = 3.74NKK188 pKa = 10.68ADD190 pKa = 3.85STPVCDD196 pKa = 3.85GTAATTPVPGTPSKK210 pKa = 10.7CRR212 pKa = 11.84CDD214 pKa = 3.58EE215 pKa = 4.04FNIDD219 pKa = 4.19VFIKK223 pKa = 10.28PDD225 pKa = 3.62PPTINKK231 pKa = 9.74SEE233 pKa = 4.11GSPITRR239 pKa = 11.84PEE241 pKa = 3.88PGGDD245 pKa = 3.46YY246 pKa = 10.71TFTASFTNNSLTSIFIDD263 pKa = 3.74SVADD267 pKa = 4.44EE268 pKa = 4.53IDD270 pKa = 3.38VGGEE274 pKa = 3.92GSFDD278 pKa = 3.55VSLDD282 pKa = 3.69LLNGATQTVDD292 pKa = 3.26PTNTATAEE300 pKa = 4.12GVYY303 pKa = 10.23LKK305 pKa = 10.72SLSASCAQPADD316 pKa = 3.42IGNGAGEE323 pKa = 4.44LVSGATYY330 pKa = 10.63SCTFTVTIVDD340 pKa = 3.46RR341 pKa = 11.84DD342 pKa = 3.82LPNDD346 pKa = 3.63QSPEE350 pKa = 3.99LYY352 pKa = 10.42KK353 pKa = 10.95DD354 pKa = 3.68VIKK357 pKa = 11.06VALTDD362 pKa = 3.6KK363 pKa = 11.05NGNPVVNGATCPADD377 pKa = 3.92LSPIAGDD384 pKa = 3.29HH385 pKa = 6.26CSVLRR390 pKa = 11.84TVDD393 pKa = 3.52VTNLPPSITVSKK405 pKa = 9.51TPSPDD410 pKa = 3.06QVLEE414 pKa = 4.09PGGDD418 pKa = 3.33VTFDD422 pKa = 3.0IVVTSTSGNYY432 pKa = 9.69DD433 pKa = 3.51DD434 pKa = 5.89PLTLTSLMDD443 pKa = 3.67TVFGDD448 pKa = 3.75LNGVGTCATGGSFSLAAPYY467 pKa = 10.35SCSFTEE473 pKa = 4.78TISGNAGDD481 pKa = 3.79VHH483 pKa = 7.0NNTVTAKK490 pKa = 9.97ATDD493 pKa = 3.69NEE495 pKa = 3.93NDD497 pKa = 3.23EE498 pKa = 4.66AQNSDD503 pKa = 3.16GATVNINDD511 pKa = 3.72VPSIITLVKK520 pKa = 8.67TADD523 pKa = 4.43PIEE526 pKa = 4.16VDD528 pKa = 3.57EE529 pKa = 5.05TGDD532 pKa = 3.64DD533 pKa = 3.47PTVFRR538 pKa = 11.84DD539 pKa = 3.0VDD541 pKa = 4.04FTFEE545 pKa = 4.08FSVDD549 pKa = 3.1AAGVDD554 pKa = 3.55DD555 pKa = 3.95VTFNSLTDD563 pKa = 3.95TIFGTLTGEE572 pKa = 4.41CLVDD576 pKa = 3.6TKK578 pKa = 10.95NGSPIGSTPLSGFVLQPGEE597 pKa = 4.45DD598 pKa = 3.61ASCTITKK605 pKa = 10.14ALQGDD610 pKa = 3.82AGDD613 pKa = 3.61VHH615 pKa = 7.69NNVATIKK622 pKa = 11.11GIDD625 pKa = 3.82EE626 pKa = 4.52DD627 pKa = 4.32AQPVMDD633 pKa = 5.29SDD635 pKa = 4.19DD636 pKa = 3.68ATVTFLDD643 pKa = 3.71TGLDD647 pKa = 3.63MEE649 pKa = 4.74QEE651 pKa = 4.44FAAKK655 pKa = 9.45MIAFVRR661 pKa = 11.84ITNGNVDD668 pKa = 3.68TASITGVTIKK678 pKa = 10.61GVNLVAGGGIPGQFEE693 pKa = 4.33VQNDD697 pKa = 3.92AGTSSYY703 pKa = 11.54NPGDD707 pKa = 3.56GPYY710 pKa = 9.83AFCSTGVDD718 pKa = 3.31ILPGDD723 pKa = 3.83TYY725 pKa = 11.69EE726 pKa = 4.9CAFTLKK732 pKa = 10.12LFPGFPPGDD741 pKa = 3.52INFSATGANGLVFTYY756 pKa = 10.51QDD758 pKa = 3.38NEE760 pKa = 4.48GNSVTTNVDD769 pKa = 3.5MNVMTIEE776 pKa = 3.87
Molecular weight: 81.33 kDa
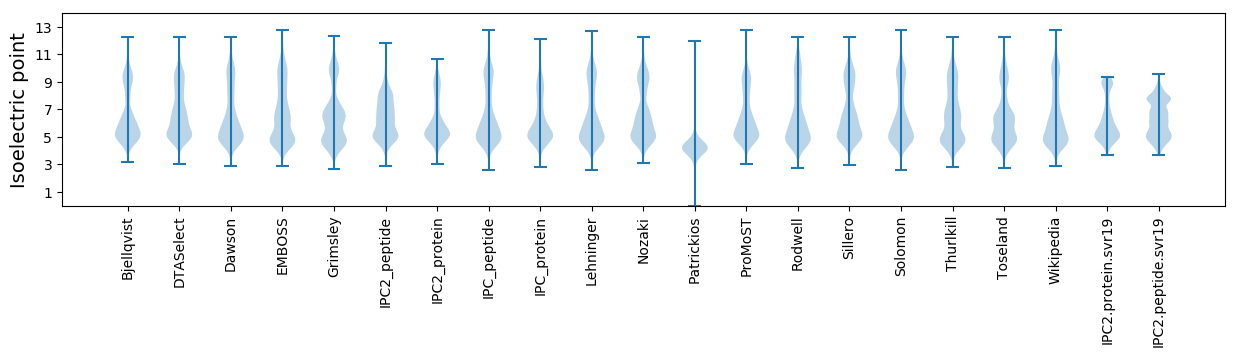
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1G8PSR4|A0A1G8PSR4_9GAMM Deoxyribose-phosphate aldolase OS=Ferrimonas sediminum OX=718193 GN=deoC PE=3 SV=1
MM1 pKa = 7.35NLKK4 pKa = 10.01FWQRR8 pKa = 11.84PPLQASPQRR17 pKa = 11.84TLGHH21 pKa = 6.49NDD23 pKa = 2.75ILILPTRR30 pKa = 11.84FGVLFLGLSLVLFLFGSNYY49 pKa = 9.41QNNLILMLAFLMVSLFSSCLLICYY73 pKa = 9.73RR74 pKa = 11.84NLAGVTLSPQPAPGTHH90 pKa = 6.69AGDD93 pKa = 3.7PARR96 pKa = 11.84FPISLSGTGHH106 pKa = 6.66SYY108 pKa = 10.8HH109 pKa = 6.42LQLGFVQGDD118 pKa = 3.54KK119 pKa = 10.94QLLHH123 pKa = 6.38TLSATPSRR131 pKa = 11.84LEE133 pKa = 3.88VAWPSRR139 pKa = 11.84RR140 pKa = 11.84RR141 pKa = 11.84GPLLPPPLIISSRR154 pKa = 11.84YY155 pKa = 8.4PLGLCRR161 pKa = 11.84VWSRR165 pKa = 11.84LALNQAAWVWPRR177 pKa = 11.84PLAGDD182 pKa = 3.97TPQAHH187 pKa = 7.39RR188 pKa = 11.84PRR190 pKa = 11.84QEE192 pKa = 3.72QGDD195 pKa = 3.82KK196 pKa = 10.76HH197 pKa = 6.22QSNSNDD203 pKa = 3.25FDD205 pKa = 4.16GLKK208 pKa = 9.69PWQRR212 pKa = 11.84GHH214 pKa = 6.04SLGRR218 pKa = 11.84VAWKK222 pKa = 10.18QLAQQRR228 pKa = 11.84GMLVKK233 pKa = 10.59QFDD236 pKa = 4.14SPGPNPEE243 pKa = 4.74LLTVDD248 pKa = 3.3NRR250 pKa = 11.84HH251 pKa = 6.12PQLEE255 pKa = 4.32HH256 pKa = 6.76HH257 pKa = 6.54LSEE260 pKa = 4.62LAGRR264 pKa = 11.84LHH266 pKa = 7.33RR267 pKa = 11.84LHH269 pKa = 5.79QQGVPVGLSTDD280 pKa = 3.64TEE282 pKa = 4.58TLMPALGRR290 pKa = 11.84THH292 pKa = 7.26LHH294 pKa = 6.07HH295 pKa = 7.23CLTLLARR302 pKa = 11.84VEE304 pKa = 4.18PHH306 pKa = 6.67ASS308 pKa = 3.09
MM1 pKa = 7.35NLKK4 pKa = 10.01FWQRR8 pKa = 11.84PPLQASPQRR17 pKa = 11.84TLGHH21 pKa = 6.49NDD23 pKa = 2.75ILILPTRR30 pKa = 11.84FGVLFLGLSLVLFLFGSNYY49 pKa = 9.41QNNLILMLAFLMVSLFSSCLLICYY73 pKa = 9.73RR74 pKa = 11.84NLAGVTLSPQPAPGTHH90 pKa = 6.69AGDD93 pKa = 3.7PARR96 pKa = 11.84FPISLSGTGHH106 pKa = 6.66SYY108 pKa = 10.8HH109 pKa = 6.42LQLGFVQGDD118 pKa = 3.54KK119 pKa = 10.94QLLHH123 pKa = 6.38TLSATPSRR131 pKa = 11.84LEE133 pKa = 3.88VAWPSRR139 pKa = 11.84RR140 pKa = 11.84RR141 pKa = 11.84GPLLPPPLIISSRR154 pKa = 11.84YY155 pKa = 8.4PLGLCRR161 pKa = 11.84VWSRR165 pKa = 11.84LALNQAAWVWPRR177 pKa = 11.84PLAGDD182 pKa = 3.97TPQAHH187 pKa = 7.39RR188 pKa = 11.84PRR190 pKa = 11.84QEE192 pKa = 3.72QGDD195 pKa = 3.82KK196 pKa = 10.76HH197 pKa = 6.22QSNSNDD203 pKa = 3.25FDD205 pKa = 4.16GLKK208 pKa = 9.69PWQRR212 pKa = 11.84GHH214 pKa = 6.04SLGRR218 pKa = 11.84VAWKK222 pKa = 10.18QLAQQRR228 pKa = 11.84GMLVKK233 pKa = 10.59QFDD236 pKa = 4.14SPGPNPEE243 pKa = 4.74LLTVDD248 pKa = 3.3NRR250 pKa = 11.84HH251 pKa = 6.12PQLEE255 pKa = 4.32HH256 pKa = 6.76HH257 pKa = 6.54LSEE260 pKa = 4.62LAGRR264 pKa = 11.84LHH266 pKa = 7.33RR267 pKa = 11.84LHH269 pKa = 5.79QQGVPVGLSTDD280 pKa = 3.64TEE282 pKa = 4.58TLMPALGRR290 pKa = 11.84THH292 pKa = 7.26LHH294 pKa = 6.07HH295 pKa = 7.23CLTLLARR302 pKa = 11.84VEE304 pKa = 4.18PHH306 pKa = 6.67ASS308 pKa = 3.09
Molecular weight: 34.39 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1277342 |
18 |
2705 |
324.3 |
35.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.819 ± 0.052 | 1.163 ± 0.016 |
5.657 ± 0.035 | 5.722 ± 0.036 |
3.66 ± 0.026 | 7.606 ± 0.035 |
2.396 ± 0.025 | 5.008 ± 0.026 |
3.908 ± 0.037 | 11.256 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.694 ± 0.02 | 3.401 ± 0.026 |
4.437 ± 0.027 | 5.274 ± 0.038 |
5.617 ± 0.033 | 6.152 ± 0.027 |
5.043 ± 0.026 | 7.029 ± 0.034 |
1.387 ± 0.017 | 2.772 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
