
Pseudomonas phage KPP25 (Bacteriophage KPP25)
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Podoviridae; Kochitakasuvirus; Pseudomonas virus KPP25
Average proteome isoelectric point is 6.31
Get precalculated fractions of proteins
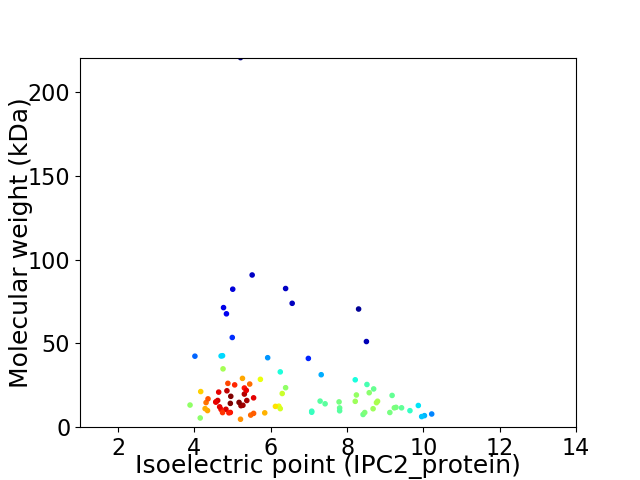
Virtual 2D-PAGE plot for 88 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|X5HZQ0|X5HZQ0_BPKP2 Uncharacterized protein OS=Pseudomonas phage KPP25 OX=1462608 PE=4 SV=1
MM1 pKa = 7.58AVTPEE6 pKa = 4.03LVYY9 pKa = 11.1DD10 pKa = 3.93GVGFVSNLVTDD21 pKa = 4.74GNQRR25 pKa = 11.84MLVLVGRR32 pKa = 11.84TQFSLNVLYY41 pKa = 11.25SNDD44 pKa = 4.87AGVTWNSTGLPPAINPGDD62 pKa = 3.57LALINGSLTDD72 pKa = 3.36GHH74 pKa = 6.72LTLTSSGYY82 pKa = 8.65TPGGQYY88 pKa = 10.79VSYY91 pKa = 10.23LFCSDD96 pKa = 3.88DD97 pKa = 5.11AGMSWSEE104 pKa = 3.69VAMPSGYY111 pKa = 10.42RR112 pKa = 11.84NFVTAGYY119 pKa = 9.29DD120 pKa = 2.99ITMGYY125 pKa = 8.98YY126 pKa = 8.36WFVVEE131 pKa = 4.67MQSSPWNRR139 pKa = 11.84AIYY142 pKa = 8.09RR143 pKa = 11.84TQDD146 pKa = 3.39LVDD149 pKa = 4.36LEE151 pKa = 5.16LIQMLPGNQEE161 pKa = 3.56GFTIQTFYY169 pKa = 11.47NNKK172 pKa = 9.67EE173 pKa = 4.2GALSWAGRR181 pKa = 11.84NAWNSSNGGNLLTGGIVGSSILTHH205 pKa = 5.39VEE207 pKa = 3.96YY208 pKa = 9.66PIVPATTYY216 pKa = 10.29FYY218 pKa = 11.21DD219 pKa = 3.6SLVICKK225 pKa = 8.97PAYY228 pKa = 9.75PNDD231 pKa = 3.55GQIYY235 pKa = 10.16LGIDD239 pKa = 3.03TSTYY243 pKa = 9.73EE244 pKa = 3.88IAVNAQQPNPAPAAYY259 pKa = 9.85FDD261 pKa = 4.32GEE263 pKa = 4.37VGIVSARR270 pKa = 11.84ITNDD274 pKa = 2.25GSNFFDD280 pKa = 4.41SYY282 pKa = 10.07FARR285 pKa = 11.84IGGTWQQTSPLPTNQQPVAFVKK307 pKa = 10.92GLDD310 pKa = 3.5DD311 pKa = 5.62LIYY314 pKa = 10.86VLGNNGILSSTVIGATFTQEE334 pKa = 3.81YY335 pKa = 9.17TGFPSSGVLSPLMGKK350 pKa = 8.77VQDD353 pKa = 4.12GLCVLFDD360 pKa = 3.33NGQIYY365 pKa = 9.56RR366 pKa = 11.84VPLLPPEE373 pKa = 4.39PPVVGKK379 pKa = 9.61FWTDD383 pKa = 2.59IEE385 pKa = 4.45YY386 pKa = 11.03CEE388 pKa = 4.29EE389 pKa = 4.01FF390 pKa = 4.07
MM1 pKa = 7.58AVTPEE6 pKa = 4.03LVYY9 pKa = 11.1DD10 pKa = 3.93GVGFVSNLVTDD21 pKa = 4.74GNQRR25 pKa = 11.84MLVLVGRR32 pKa = 11.84TQFSLNVLYY41 pKa = 11.25SNDD44 pKa = 4.87AGVTWNSTGLPPAINPGDD62 pKa = 3.57LALINGSLTDD72 pKa = 3.36GHH74 pKa = 6.72LTLTSSGYY82 pKa = 8.65TPGGQYY88 pKa = 10.79VSYY91 pKa = 10.23LFCSDD96 pKa = 3.88DD97 pKa = 5.11AGMSWSEE104 pKa = 3.69VAMPSGYY111 pKa = 10.42RR112 pKa = 11.84NFVTAGYY119 pKa = 9.29DD120 pKa = 2.99ITMGYY125 pKa = 8.98YY126 pKa = 8.36WFVVEE131 pKa = 4.67MQSSPWNRR139 pKa = 11.84AIYY142 pKa = 8.09RR143 pKa = 11.84TQDD146 pKa = 3.39LVDD149 pKa = 4.36LEE151 pKa = 5.16LIQMLPGNQEE161 pKa = 3.56GFTIQTFYY169 pKa = 11.47NNKK172 pKa = 9.67EE173 pKa = 4.2GALSWAGRR181 pKa = 11.84NAWNSSNGGNLLTGGIVGSSILTHH205 pKa = 5.39VEE207 pKa = 3.96YY208 pKa = 9.66PIVPATTYY216 pKa = 10.29FYY218 pKa = 11.21DD219 pKa = 3.6SLVICKK225 pKa = 8.97PAYY228 pKa = 9.75PNDD231 pKa = 3.55GQIYY235 pKa = 10.16LGIDD239 pKa = 3.03TSTYY243 pKa = 9.73EE244 pKa = 3.88IAVNAQQPNPAPAAYY259 pKa = 9.85FDD261 pKa = 4.32GEE263 pKa = 4.37VGIVSARR270 pKa = 11.84ITNDD274 pKa = 2.25GSNFFDD280 pKa = 4.41SYY282 pKa = 10.07FARR285 pKa = 11.84IGGTWQQTSPLPTNQQPVAFVKK307 pKa = 10.92GLDD310 pKa = 3.5DD311 pKa = 5.62LIYY314 pKa = 10.86VLGNNGILSSTVIGATFTQEE334 pKa = 3.81YY335 pKa = 9.17TGFPSSGVLSPLMGKK350 pKa = 8.77VQDD353 pKa = 4.12GLCVLFDD360 pKa = 3.33NGQIYY365 pKa = 9.56RR366 pKa = 11.84VPLLPPEE373 pKa = 4.39PPVVGKK379 pKa = 9.61FWTDD383 pKa = 2.59IEE385 pKa = 4.45YY386 pKa = 11.03CEE388 pKa = 4.29EE389 pKa = 4.01FF390 pKa = 4.07
Molecular weight: 42.4 kDa
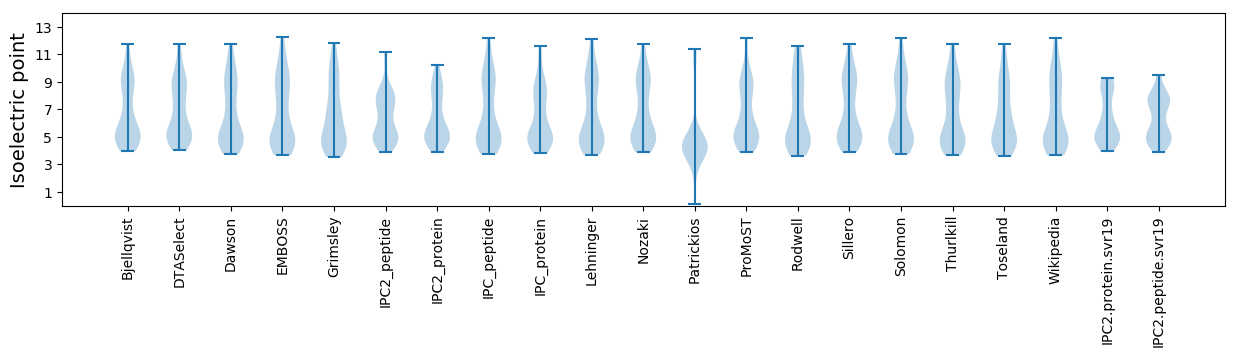
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|X5I375|X5I375_BPKP2 Uncharacterized protein OS=Pseudomonas phage KPP25 OX=1462608 PE=4 SV=1
MM1 pKa = 7.04STFAALKK8 pKa = 9.7NLGSAIVSDD17 pKa = 3.98IAAGARR23 pKa = 11.84ATASATSSAYY33 pKa = 9.78EE34 pKa = 4.09SVSAKK39 pKa = 9.55VAHH42 pKa = 6.62KK43 pKa = 10.32RR44 pKa = 11.84GEE46 pKa = 3.84RR47 pKa = 11.84QIINEE52 pKa = 4.04FRR54 pKa = 11.84NMLNEE59 pKa = 4.17HH60 pKa = 6.7PGEE63 pKa = 4.04LLAAMRR69 pKa = 11.84SIQDD73 pKa = 3.45QLAQPAKK80 pKa = 9.6PARR83 pKa = 11.84KK84 pKa = 8.55PRR86 pKa = 11.84ARR88 pKa = 11.84KK89 pKa = 8.7AAAQQ93 pKa = 3.42
MM1 pKa = 7.04STFAALKK8 pKa = 9.7NLGSAIVSDD17 pKa = 3.98IAAGARR23 pKa = 11.84ATASATSSAYY33 pKa = 9.78EE34 pKa = 4.09SVSAKK39 pKa = 9.55VAHH42 pKa = 6.62KK43 pKa = 10.32RR44 pKa = 11.84GEE46 pKa = 3.84RR47 pKa = 11.84QIINEE52 pKa = 4.04FRR54 pKa = 11.84NMLNEE59 pKa = 4.17HH60 pKa = 6.7PGEE63 pKa = 4.04LLAAMRR69 pKa = 11.84SIQDD73 pKa = 3.45QLAQPAKK80 pKa = 9.6PARR83 pKa = 11.84KK84 pKa = 8.55PRR86 pKa = 11.84ARR88 pKa = 11.84KK89 pKa = 8.7AAAQQ93 pKa = 3.42
Molecular weight: 9.87 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
19981 |
42 |
2009 |
227.1 |
25.13 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.61 ± 0.453 | 1.031 ± 0.168 |
6.516 ± 0.209 | 6.191 ± 0.329 |
3.438 ± 0.198 | 6.831 ± 0.36 |
1.677 ± 0.153 | 4.98 ± 0.204 |
4.529 ± 0.39 | 8.568 ± 0.264 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.698 ± 0.131 | 3.939 ± 0.191 |
5.055 ± 0.199 | 4.244 ± 0.294 |
6.676 ± 0.369 | 5.71 ± 0.209 |
6.256 ± 0.226 | 6.746 ± 0.23 |
1.316 ± 0.104 | 2.988 ± 0.199 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
