
Gammaproteobacteria bacterium
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; unclassified Gammaproteobacteria
Average proteome isoelectric point is 6.46
Get precalculated fractions of proteins
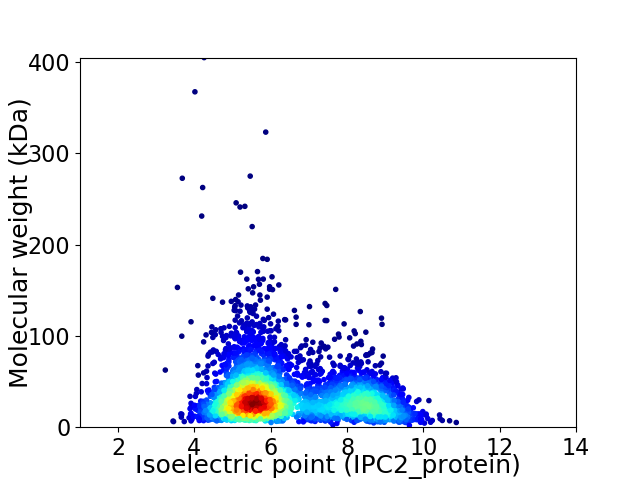
Virtual 2D-PAGE plot for 3997 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Q5VCC0|A0A4Q5VCC0_9GAMM Flavin reductase OS=Gammaproteobacteria bacterium OX=1913989 GN=EOO52_07800 PE=4 SV=1
LL1 pKa = 6.68GTDD4 pKa = 3.62GDD6 pKa = 4.06AGTTVTYY13 pKa = 11.01SLTDD17 pKa = 3.3NAGGRR22 pKa = 11.84FAINSTTGVITVANGSLLDD41 pKa = 3.89YY42 pKa = 11.01EE43 pKa = 5.05SATSHH48 pKa = 5.83NVTVLATSSDD58 pKa = 3.13GSTANRR64 pKa = 11.84AFSISVTNVNDD75 pKa = 3.76APVATAATYY84 pKa = 9.03TGNEE88 pKa = 3.84DD89 pKa = 3.31AAFITVNLAGTDD101 pKa = 3.29VDD103 pKa = 3.99GTIASLKK110 pKa = 10.18VSSLPTAAQGVLYY123 pKa = 10.53LADD126 pKa = 3.62GVTAVTTAMNLTPAQAAALKK146 pKa = 8.9FVPKK150 pKa = 10.5LNYY153 pKa = 10.23NGEE156 pKa = 3.83FDD158 pKa = 3.81IAFTATDD165 pKa = 3.11NSGAVSSPANAHH177 pKa = 4.66VVVNAVNDD185 pKa = 3.8APIANNDD192 pKa = 3.45PTASSSVGVGLLSQYY207 pKa = 9.86FAYY210 pKa = 10.83NEE212 pKa = 4.22GVNGGNLTSIAQMNAFISGRR232 pKa = 11.84TPDD235 pKa = 3.2ATFVAKK241 pKa = 9.63TFNYY245 pKa = 10.4GSDD248 pKa = 3.16NLFSNDD254 pKa = 3.5LGRR257 pKa = 11.84GTNLQSFLGSDD268 pKa = 3.59AASLSTDD275 pKa = 3.79PGDD278 pKa = 3.68STDD281 pKa = 4.6AIIRR285 pKa = 11.84MYY287 pKa = 10.96GAVEE291 pKa = 4.2LAAGTYY297 pKa = 8.95NFQVRR302 pKa = 11.84GDD304 pKa = 3.96DD305 pKa = 4.06GYY307 pKa = 11.28QIKK310 pKa = 10.62VDD312 pKa = 3.86GKK314 pKa = 10.41VVALVDD320 pKa = 5.48AIQSPTGTVHH330 pKa = 6.8SQFTLADD337 pKa = 3.88SGLHH341 pKa = 5.87TIEE344 pKa = 4.46ILYY347 pKa = 9.34WDD349 pKa = 3.64QGGQAVFKK357 pKa = 10.98IEE359 pKa = 4.57LSDD362 pKa = 3.91NNGVSYY368 pKa = 11.27NLLSSKK374 pKa = 8.51PTSYY378 pKa = 11.07NAVYY382 pKa = 10.54SLNEE386 pKa = 3.81DD387 pKa = 4.4TIWSVPVSTLLANDD401 pKa = 3.69TDD403 pKa = 3.86VDD405 pKa = 4.09GDD407 pKa = 3.88TLSIVSVQNALHH419 pKa = 5.78GTVQIVGSNVVFTPEE434 pKa = 3.78ANYY437 pKa = 10.84SGIASYY443 pKa = 9.69TYY445 pKa = 9.98TISDD449 pKa = 3.71GKK451 pKa = 11.05GGTSTAKK458 pKa = 10.53VNIDD462 pKa = 3.27IQPLNDD468 pKa = 3.8APLAVNDD475 pKa = 4.24NLTATEE481 pKa = 4.59DD482 pKa = 3.37ASITYY487 pKa = 8.04TAAQLLGNDD496 pKa = 3.61SDD498 pKa = 4.06VDD500 pKa = 3.92SSAFTISSVTSGTGGTAVLNANGTVTFTPTANFNGNATFTYY541 pKa = 10.29RR542 pKa = 11.84ATDD545 pKa = 3.99GSAISAPATVTVAVASVNDD564 pKa = 3.99APSGTSSSVGIIEE577 pKa = 4.72DD578 pKa = 3.44QSHH581 pKa = 6.14TLTVANFGFTDD592 pKa = 3.5TADD595 pKa = 3.87NNSLLAVRR603 pKa = 11.84ISTLPTSGTLYY614 pKa = 10.35LNNVAVTAGTYY625 pKa = 10.13ISVADD630 pKa = 3.89ISAGKK635 pKa = 10.18LIYY638 pKa = 10.49LPSNNDD644 pKa = 2.69SGTARR649 pKa = 11.84TFTFQVQDD657 pKa = 4.05DD658 pKa = 4.26GGTANGGVNLDD669 pKa = 3.64ATPNTLTFNIADD681 pKa = 3.76VPTTTDD687 pKa = 2.64KK688 pKa = 11.22TGNTNANTFAVLANSGNFSILDD710 pKa = 3.51SGGTDD715 pKa = 3.43SISVTTATGAVFNTLNFEE733 pKa = 4.42HH734 pKa = 6.62VGNNLEE740 pKa = 4.22FTQATAAGSANVSVINQYY758 pKa = 9.84VAANTIEE765 pKa = 4.17TLSFSSGGDD774 pKa = 3.27YY775 pKa = 11.25AGFQVGTAATTTYY788 pKa = 10.46TLNTTNTGTSGNDD801 pKa = 3.08IIVGTTADD809 pKa = 3.79DD810 pKa = 4.4TISGGGGAGRR820 pKa = 11.84DD821 pKa = 3.67MLFGGAGNDD830 pKa = 3.81TITAAGTGNHH840 pKa = 6.36LLVGEE845 pKa = 5.15LGNDD849 pKa = 3.66TLIGGAGSDD858 pKa = 3.29WLIGGKK864 pKa = 10.65GNDD867 pKa = 3.98SLTGGAGVDD876 pKa = 3.42TFVWQLADD884 pKa = 3.58KK885 pKa = 9.09GTTAAPAVDD894 pKa = 3.98TVNDD898 pKa = 3.64SDD900 pKa = 6.1GSDD903 pKa = 3.41KK904 pKa = 11.56LNLSDD909 pKa = 5.58LLQGEE914 pKa = 4.32NSGNLTNYY922 pKa = 10.42LHH924 pKa = 6.06FTSDD928 pKa = 3.17GTTTTVSISSTGAFDD943 pKa = 3.62GSNYY947 pKa = 8.18GTATDD952 pKa = 3.46QTIILTNVNLTGGDD966 pKa = 3.35AAIINLLKK974 pKa = 10.23TNNNLITDD982 pKa = 4.29
LL1 pKa = 6.68GTDD4 pKa = 3.62GDD6 pKa = 4.06AGTTVTYY13 pKa = 11.01SLTDD17 pKa = 3.3NAGGRR22 pKa = 11.84FAINSTTGVITVANGSLLDD41 pKa = 3.89YY42 pKa = 11.01EE43 pKa = 5.05SATSHH48 pKa = 5.83NVTVLATSSDD58 pKa = 3.13GSTANRR64 pKa = 11.84AFSISVTNVNDD75 pKa = 3.76APVATAATYY84 pKa = 9.03TGNEE88 pKa = 3.84DD89 pKa = 3.31AAFITVNLAGTDD101 pKa = 3.29VDD103 pKa = 3.99GTIASLKK110 pKa = 10.18VSSLPTAAQGVLYY123 pKa = 10.53LADD126 pKa = 3.62GVTAVTTAMNLTPAQAAALKK146 pKa = 8.9FVPKK150 pKa = 10.5LNYY153 pKa = 10.23NGEE156 pKa = 3.83FDD158 pKa = 3.81IAFTATDD165 pKa = 3.11NSGAVSSPANAHH177 pKa = 4.66VVVNAVNDD185 pKa = 3.8APIANNDD192 pKa = 3.45PTASSSVGVGLLSQYY207 pKa = 9.86FAYY210 pKa = 10.83NEE212 pKa = 4.22GVNGGNLTSIAQMNAFISGRR232 pKa = 11.84TPDD235 pKa = 3.2ATFVAKK241 pKa = 9.63TFNYY245 pKa = 10.4GSDD248 pKa = 3.16NLFSNDD254 pKa = 3.5LGRR257 pKa = 11.84GTNLQSFLGSDD268 pKa = 3.59AASLSTDD275 pKa = 3.79PGDD278 pKa = 3.68STDD281 pKa = 4.6AIIRR285 pKa = 11.84MYY287 pKa = 10.96GAVEE291 pKa = 4.2LAAGTYY297 pKa = 8.95NFQVRR302 pKa = 11.84GDD304 pKa = 3.96DD305 pKa = 4.06GYY307 pKa = 11.28QIKK310 pKa = 10.62VDD312 pKa = 3.86GKK314 pKa = 10.41VVALVDD320 pKa = 5.48AIQSPTGTVHH330 pKa = 6.8SQFTLADD337 pKa = 3.88SGLHH341 pKa = 5.87TIEE344 pKa = 4.46ILYY347 pKa = 9.34WDD349 pKa = 3.64QGGQAVFKK357 pKa = 10.98IEE359 pKa = 4.57LSDD362 pKa = 3.91NNGVSYY368 pKa = 11.27NLLSSKK374 pKa = 8.51PTSYY378 pKa = 11.07NAVYY382 pKa = 10.54SLNEE386 pKa = 3.81DD387 pKa = 4.4TIWSVPVSTLLANDD401 pKa = 3.69TDD403 pKa = 3.86VDD405 pKa = 4.09GDD407 pKa = 3.88TLSIVSVQNALHH419 pKa = 5.78GTVQIVGSNVVFTPEE434 pKa = 3.78ANYY437 pKa = 10.84SGIASYY443 pKa = 9.69TYY445 pKa = 9.98TISDD449 pKa = 3.71GKK451 pKa = 11.05GGTSTAKK458 pKa = 10.53VNIDD462 pKa = 3.27IQPLNDD468 pKa = 3.8APLAVNDD475 pKa = 4.24NLTATEE481 pKa = 4.59DD482 pKa = 3.37ASITYY487 pKa = 8.04TAAQLLGNDD496 pKa = 3.61SDD498 pKa = 4.06VDD500 pKa = 3.92SSAFTISSVTSGTGGTAVLNANGTVTFTPTANFNGNATFTYY541 pKa = 10.29RR542 pKa = 11.84ATDD545 pKa = 3.99GSAISAPATVTVAVASVNDD564 pKa = 3.99APSGTSSSVGIIEE577 pKa = 4.72DD578 pKa = 3.44QSHH581 pKa = 6.14TLTVANFGFTDD592 pKa = 3.5TADD595 pKa = 3.87NNSLLAVRR603 pKa = 11.84ISTLPTSGTLYY614 pKa = 10.35LNNVAVTAGTYY625 pKa = 10.13ISVADD630 pKa = 3.89ISAGKK635 pKa = 10.18LIYY638 pKa = 10.49LPSNNDD644 pKa = 2.69SGTARR649 pKa = 11.84TFTFQVQDD657 pKa = 4.05DD658 pKa = 4.26GGTANGGVNLDD669 pKa = 3.64ATPNTLTFNIADD681 pKa = 3.76VPTTTDD687 pKa = 2.64KK688 pKa = 11.22TGNTNANTFAVLANSGNFSILDD710 pKa = 3.51SGGTDD715 pKa = 3.43SISVTTATGAVFNTLNFEE733 pKa = 4.42HH734 pKa = 6.62VGNNLEE740 pKa = 4.22FTQATAAGSANVSVINQYY758 pKa = 9.84VAANTIEE765 pKa = 4.17TLSFSSGGDD774 pKa = 3.27YY775 pKa = 11.25AGFQVGTAATTTYY788 pKa = 10.46TLNTTNTGTSGNDD801 pKa = 3.08IIVGTTADD809 pKa = 3.79DD810 pKa = 4.4TISGGGGAGRR820 pKa = 11.84DD821 pKa = 3.67MLFGGAGNDD830 pKa = 3.81TITAAGTGNHH840 pKa = 6.36LLVGEE845 pKa = 5.15LGNDD849 pKa = 3.66TLIGGAGSDD858 pKa = 3.29WLIGGKK864 pKa = 10.65GNDD867 pKa = 3.98SLTGGAGVDD876 pKa = 3.42TFVWQLADD884 pKa = 3.58KK885 pKa = 9.09GTTAAPAVDD894 pKa = 3.98TVNDD898 pKa = 3.64SDD900 pKa = 6.1GSDD903 pKa = 3.41KK904 pKa = 11.56LNLSDD909 pKa = 5.58LLQGEE914 pKa = 4.32NSGNLTNYY922 pKa = 10.42LHH924 pKa = 6.06FTSDD928 pKa = 3.17GTTTTVSISSTGAFDD943 pKa = 3.62GSNYY947 pKa = 8.18GTATDD952 pKa = 3.46QTIILTNVNLTGGDD966 pKa = 3.35AAIINLLKK974 pKa = 10.23TNNNLITDD982 pKa = 4.29
Molecular weight: 99.68 kDa
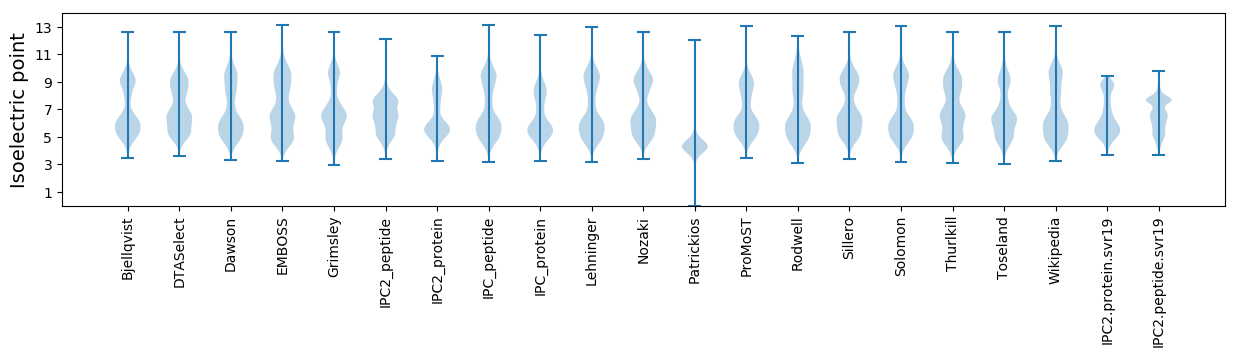
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Q5VHC1|A0A4Q5VHC1_9GAMM tRNA-specific adenosine deaminase OS=Gammaproteobacteria bacterium OX=1913989 GN=tadA PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSTLKK11 pKa = 10.47RR12 pKa = 11.84ARR14 pKa = 11.84NHH16 pKa = 5.29GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.35NGRR28 pKa = 11.84LVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.0GRR39 pKa = 11.84KK40 pKa = 8.22EE41 pKa = 3.7LTVV44 pKa = 3.18
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSTLKK11 pKa = 10.47RR12 pKa = 11.84ARR14 pKa = 11.84NHH16 pKa = 5.29GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.35NGRR28 pKa = 11.84LVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.0GRR39 pKa = 11.84KK40 pKa = 8.22EE41 pKa = 3.7LTVV44 pKa = 3.18
Molecular weight: 5.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1350151 |
18 |
3945 |
337.8 |
37.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.032 ± 0.043 | 0.956 ± 0.015 |
5.427 ± 0.031 | 5.792 ± 0.041 |
4.236 ± 0.024 | 6.892 ± 0.045 |
2.114 ± 0.019 | 6.446 ± 0.033 |
5.377 ± 0.033 | 10.102 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.18 ± 0.017 | 4.735 ± 0.037 |
4.141 ± 0.026 | 4.029 ± 0.027 |
4.806 ± 0.033 | 6.99 ± 0.051 |
5.592 ± 0.041 | 6.785 ± 0.034 |
1.309 ± 0.017 | 3.058 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
