
Dechloromonas sp.
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Rhodocyclales; Azonexaceae; Dechloromonas; unclassified Dechloromonas
Average proteome isoelectric point is 6.82
Get precalculated fractions of proteins
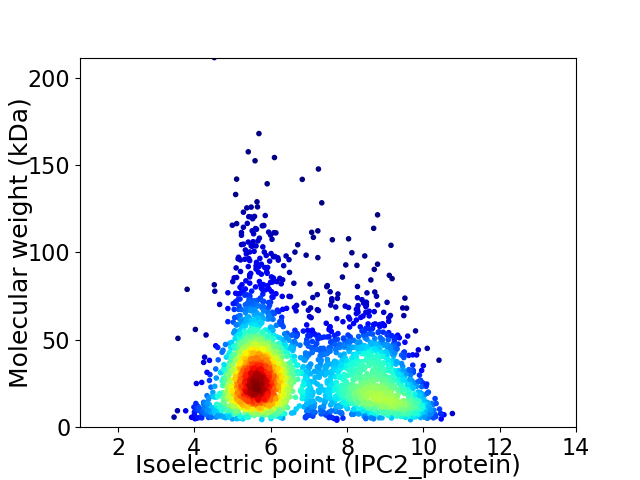
Virtual 2D-PAGE plot for 2873 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
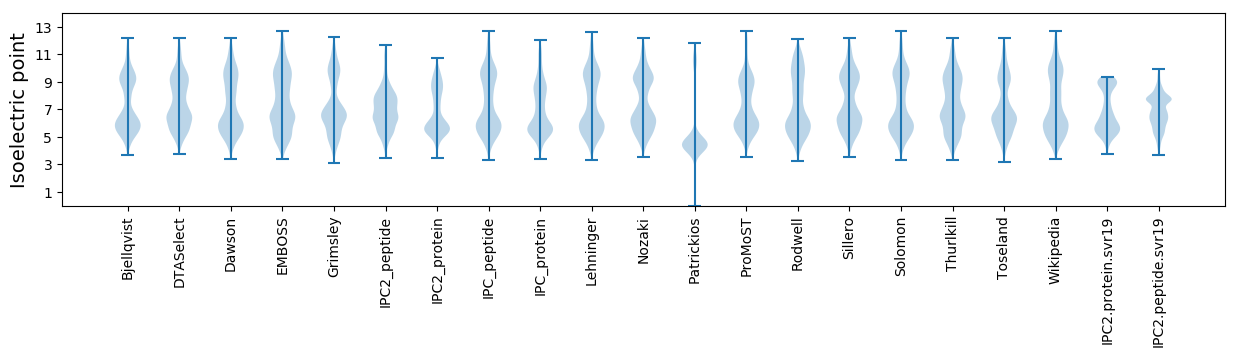
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6N1XG58|A0A6N1XG58_9RHOO Hydrolase or metal-binding protein OS=Dechloromonas sp. OX=1917218 GN=HT580_10515 PE=4 SV=1
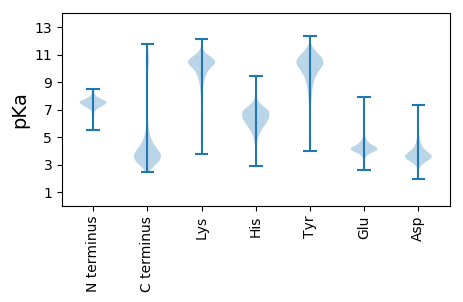
MM1 pKa = 7.02NTLLDD6 pKa = 3.82ASISGTSPFAVTLTGLPAGTVVTGMMMTMVGGAPVWSAQGTGDD49 pKa = 3.96NNALQALLNGITVTAPANANSNTGPFGFSATLTTYY84 pKa = 11.22DD85 pKa = 4.33DD86 pKa = 3.69GGGRR90 pKa = 11.84HH91 pKa = 6.64DD92 pKa = 4.69SNLTVSLPVTPVSDD106 pKa = 4.73PIVLVATDD114 pKa = 4.07SNVSEE119 pKa = 5.37DD120 pKa = 3.53SLAPISIALSNVADD134 pKa = 4.09GANTNVVGGKK144 pKa = 9.96VYY146 pKa = 10.64VRR148 pKa = 11.84LDD150 pKa = 3.55EE151 pKa = 4.92SGMEE155 pKa = 3.94AAGGVLRR162 pKa = 11.84FNGNPVAATSVSGVSGIADD181 pKa = 3.17GTYY184 pKa = 9.77FVLNGVSNGATLNLTYY200 pKa = 10.6QPAANASGTVGYY212 pKa = 7.91TAFVQNQEE220 pKa = 3.6TGAANVATSSTSGSFDD236 pKa = 3.19INKK239 pKa = 9.38VNDD242 pKa = 3.97GVTLSAGNVTGAEE255 pKa = 4.06DD256 pKa = 3.73QAVQLAISAALIDD269 pKa = 3.73SGEE272 pKa = 4.41RR273 pKa = 11.84IQSVTLSHH281 pKa = 6.61VPDD284 pKa = 4.72GFQVYY289 pKa = 9.41YY290 pKa = 11.26GNGAPGTAATNMGGGVWSIPASAYY314 pKa = 9.91AVPAYY319 pKa = 9.56ISLVPPPNWSGTVSGVTADD338 pKa = 3.17VWSGEE343 pKa = 4.18TGLDD347 pKa = 3.38QTVTSANFDD356 pKa = 3.52VTFNGVADD364 pKa = 5.79GILISPTLSFGNEE377 pKa = 3.98GQVVPLNLNSAMPDD391 pKa = 3.02MDD393 pKa = 3.66GSEE396 pKa = 4.08TAIITVKK403 pKa = 10.85GLGSFAAFYY412 pKa = 10.52AGSSLLSATYY422 pKa = 10.6DD423 pKa = 3.49SGLDD427 pKa = 3.39TYY429 pKa = 9.61TLSGLTPAQVSSLGVIQKK447 pKa = 10.41DD448 pKa = 3.71GSFDD452 pKa = 4.08LVITAQTADD461 pKa = 3.66SPGGNLSAVVNASMHH476 pKa = 5.98VDD478 pKa = 3.77INPVAATTGDD488 pKa = 3.63DD489 pKa = 3.19RR490 pKa = 11.84LLYY493 pKa = 10.64DD494 pKa = 4.06GSALNGLVGSTPSNSAA510 pKa = 3.0
MM1 pKa = 7.02NTLLDD6 pKa = 3.82ASISGTSPFAVTLTGLPAGTVVTGMMMTMVGGAPVWSAQGTGDD49 pKa = 3.96NNALQALLNGITVTAPANANSNTGPFGFSATLTTYY84 pKa = 11.22DD85 pKa = 4.33DD86 pKa = 3.69GGGRR90 pKa = 11.84HH91 pKa = 6.64DD92 pKa = 4.69SNLTVSLPVTPVSDD106 pKa = 4.73PIVLVATDD114 pKa = 4.07SNVSEE119 pKa = 5.37DD120 pKa = 3.53SLAPISIALSNVADD134 pKa = 4.09GANTNVVGGKK144 pKa = 9.96VYY146 pKa = 10.64VRR148 pKa = 11.84LDD150 pKa = 3.55EE151 pKa = 4.92SGMEE155 pKa = 3.94AAGGVLRR162 pKa = 11.84FNGNPVAATSVSGVSGIADD181 pKa = 3.17GTYY184 pKa = 9.77FVLNGVSNGATLNLTYY200 pKa = 10.6QPAANASGTVGYY212 pKa = 7.91TAFVQNQEE220 pKa = 3.6TGAANVATSSTSGSFDD236 pKa = 3.19INKK239 pKa = 9.38VNDD242 pKa = 3.97GVTLSAGNVTGAEE255 pKa = 4.06DD256 pKa = 3.73QAVQLAISAALIDD269 pKa = 3.73SGEE272 pKa = 4.41RR273 pKa = 11.84IQSVTLSHH281 pKa = 6.61VPDD284 pKa = 4.72GFQVYY289 pKa = 9.41YY290 pKa = 11.26GNGAPGTAATNMGGGVWSIPASAYY314 pKa = 9.91AVPAYY319 pKa = 9.56ISLVPPPNWSGTVSGVTADD338 pKa = 3.17VWSGEE343 pKa = 4.18TGLDD347 pKa = 3.38QTVTSANFDD356 pKa = 3.52VTFNGVADD364 pKa = 5.79GILISPTLSFGNEE377 pKa = 3.98GQVVPLNLNSAMPDD391 pKa = 3.02MDD393 pKa = 3.66GSEE396 pKa = 4.08TAIITVKK403 pKa = 10.85GLGSFAAFYY412 pKa = 10.52AGSSLLSATYY422 pKa = 10.6DD423 pKa = 3.49SGLDD427 pKa = 3.39TYY429 pKa = 9.61TLSGLTPAQVSSLGVIQKK447 pKa = 10.41DD448 pKa = 3.71GSFDD452 pKa = 4.08LVITAQTADD461 pKa = 3.66SPGGNLSAVVNASMHH476 pKa = 5.98VDD478 pKa = 3.77INPVAATTGDD488 pKa = 3.63DD489 pKa = 3.19RR490 pKa = 11.84LLYY493 pKa = 10.64DD494 pKa = 4.06GSALNGLVGSTPSNSAA510 pKa = 3.0
Molecular weight: 50.86 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6N1XEQ9|A0A6N1XEQ9_9RHOO YceI family protein OS=Dechloromonas sp. OX=1917218 GN=HT580_07725 PE=4 SV=1
MM1 pKa = 6.82STWIEE6 pKa = 3.85SSIPFVAIFVMLIVGTDD23 pKa = 3.2LRR25 pKa = 11.84LADD28 pKa = 4.05FQRR31 pKa = 11.84VRR33 pKa = 11.84RR34 pKa = 11.84YY35 pKa = 9.74LVLVPGIVCGQWLLLIGVAGLVGRR59 pKa = 11.84LLDD62 pKa = 4.78LPPGMTGGAILLAAAPVAAMSGFYY86 pKa = 9.06TQLAGGHH93 pKa = 6.63LALAVTVAAVSNLLAPIVTPVAASLGFWLFLDD125 pKa = 3.62SDD127 pKa = 4.18KK128 pKa = 11.8AMEE131 pKa = 4.41LPMLKK136 pKa = 10.01VALQAIVGLLLPLLAGMLLRR156 pKa = 11.84HH157 pKa = 6.27CAPRR161 pKa = 11.84WVTRR165 pKa = 11.84AAXLQGIAIGAVIALLGAVIVDD187 pKa = 3.22QXRR190 pKa = 11.84SNRR193 pKa = 11.84EE194 pKa = 3.45QFVLLFGVSLLFTLATLAAGWVALRR219 pKa = 11.84LPACSTEE226 pKa = 3.85DD227 pKa = 2.93RR228 pKa = 11.84XRR230 pKa = 11.84AALGFPARR238 pKa = 11.84NVAVATLAATSVLGKK253 pKa = 10.24AAVVTFIAVLFTTQLTLLLSLALWLRR279 pKa = 11.84KK280 pKa = 9.2RR281 pKa = 11.84AA282 pKa = 3.58
MM1 pKa = 6.82STWIEE6 pKa = 3.85SSIPFVAIFVMLIVGTDD23 pKa = 3.2LRR25 pKa = 11.84LADD28 pKa = 4.05FQRR31 pKa = 11.84VRR33 pKa = 11.84RR34 pKa = 11.84YY35 pKa = 9.74LVLVPGIVCGQWLLLIGVAGLVGRR59 pKa = 11.84LLDD62 pKa = 4.78LPPGMTGGAILLAAAPVAAMSGFYY86 pKa = 9.06TQLAGGHH93 pKa = 6.63LALAVTVAAVSNLLAPIVTPVAASLGFWLFLDD125 pKa = 3.62SDD127 pKa = 4.18KK128 pKa = 11.8AMEE131 pKa = 4.41LPMLKK136 pKa = 10.01VALQAIVGLLLPLLAGMLLRR156 pKa = 11.84HH157 pKa = 6.27CAPRR161 pKa = 11.84WVTRR165 pKa = 11.84AAXLQGIAIGAVIALLGAVIVDD187 pKa = 3.22QXRR190 pKa = 11.84SNRR193 pKa = 11.84EE194 pKa = 3.45QFVLLFGVSLLFTLATLAAGWVALRR219 pKa = 11.84LPACSTEE226 pKa = 3.85DD227 pKa = 2.93RR228 pKa = 11.84XRR230 pKa = 11.84AALGFPARR238 pKa = 11.84NVAVATLAATSVLGKK253 pKa = 10.24AAVVTFIAVLFTTQLTLLLSLALWLRR279 pKa = 11.84KK280 pKa = 9.2RR281 pKa = 11.84AA282 pKa = 3.58
Molecular weight: 29.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
878941 |
37 |
1957 |
305.9 |
33.3 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.749 ± 0.07 | 1.092 ± 0.018 |
5.284 ± 0.031 | 5.706 ± 0.042 |
3.799 ± 0.032 | 8.046 ± 0.044 |
2.215 ± 0.021 | 5.139 ± 0.029 |
3.825 ± 0.041 | 10.723 ± 0.061 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.429 ± 0.024 | 2.957 ± 0.029 |
5.016 ± 0.031 | 3.573 ± 0.025 |
6.826 ± 0.045 | 5.174 ± 0.032 |
4.893 ± 0.033 | 7.171 ± 0.041 |
1.414 ± 0.021 | 2.396 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
