
Scytonema millei VB511283
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; Nostocales; Scytonemataceae; Scytonema; Scytonema millei
Average proteome isoelectric point is 6.5
Get precalculated fractions of proteins
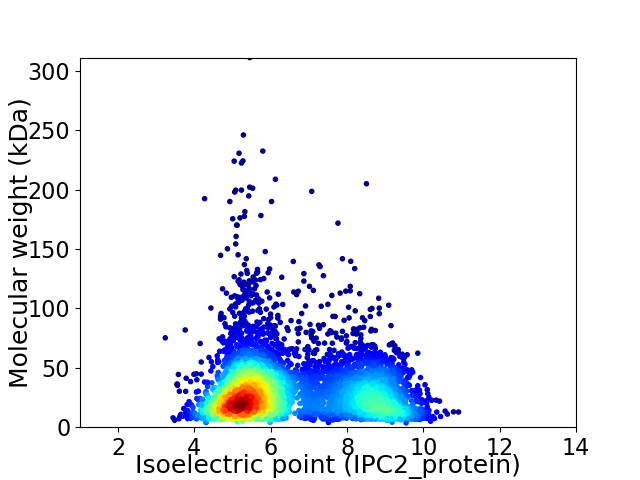
Virtual 2D-PAGE plot for 4950 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
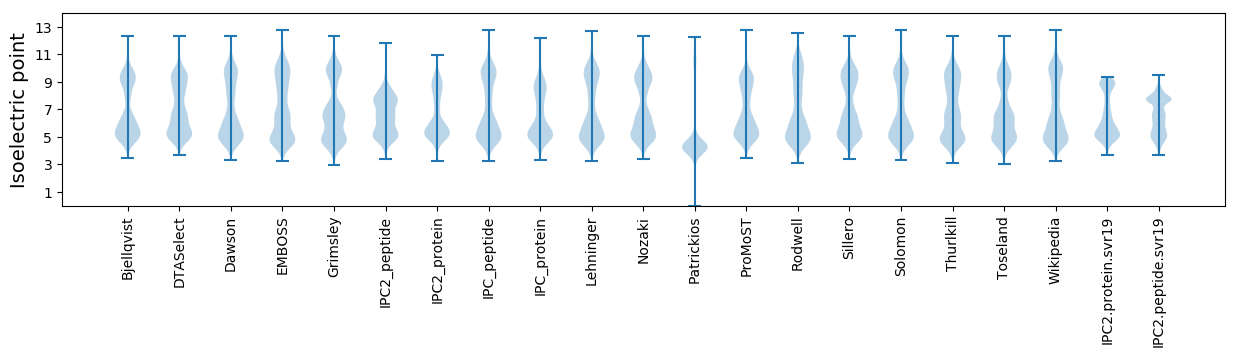
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1D2PE31|A0A1D2PE31_9CYAN FAD-dependent oxidoreductase OS=Scytonema millei VB511283 OX=1245923 GN=QH73_021535 PE=4 SV=1
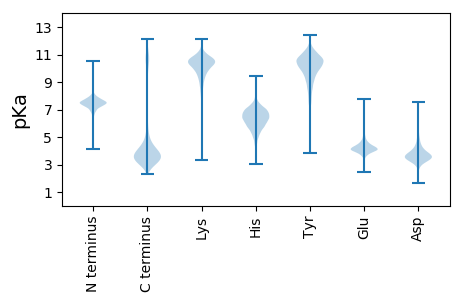
MM1 pKa = 7.31TRR3 pKa = 11.84FDD5 pKa = 4.36GDD7 pKa = 4.14SNNNTLNGTAGDD19 pKa = 4.14DD20 pKa = 3.51EE21 pKa = 5.66LYY23 pKa = 10.79GYY25 pKa = 10.31DD26 pKa = 4.26GNDD29 pKa = 2.89ILFGGDD35 pKa = 3.78GNDD38 pKa = 3.32TDD40 pKa = 4.66KK41 pKa = 11.5LVGGSGDD48 pKa = 4.54DD49 pKa = 3.48ILIGSIGKK57 pKa = 9.44DD58 pKa = 2.83LFIFNDD64 pKa = 3.23YY65 pKa = 10.08EE66 pKa = 4.83YY67 pKa = 11.18NVSIYY72 pKa = 10.65KK73 pKa = 10.08DD74 pKa = 3.16AYY76 pKa = 9.39NNYY79 pKa = 9.65HH80 pKa = 5.54VFSSEE85 pKa = 3.49GHH87 pKa = 6.75DD88 pKa = 3.32IITNFSEE95 pKa = 4.61AEE97 pKa = 3.63GDD99 pKa = 3.6KK100 pKa = 10.61IYY102 pKa = 11.31VLDD105 pKa = 4.74PQPADD110 pKa = 4.0PINRR114 pKa = 11.84IKK116 pKa = 10.72PIDD119 pKa = 4.3PIIYY123 pKa = 9.82RR124 pKa = 11.84GSADD128 pKa = 3.75NIKK131 pKa = 9.73TILGANTSQYY141 pKa = 9.88TLSNSSLMLEE151 pKa = 4.36SNILQNPLYY160 pKa = 10.93NNLSNLGADD169 pKa = 3.37TSGFGLL175 pKa = 4.02
MM1 pKa = 7.31TRR3 pKa = 11.84FDD5 pKa = 4.36GDD7 pKa = 4.14SNNNTLNGTAGDD19 pKa = 4.14DD20 pKa = 3.51EE21 pKa = 5.66LYY23 pKa = 10.79GYY25 pKa = 10.31DD26 pKa = 4.26GNDD29 pKa = 2.89ILFGGDD35 pKa = 3.78GNDD38 pKa = 3.32TDD40 pKa = 4.66KK41 pKa = 11.5LVGGSGDD48 pKa = 4.54DD49 pKa = 3.48ILIGSIGKK57 pKa = 9.44DD58 pKa = 2.83LFIFNDD64 pKa = 3.23YY65 pKa = 10.08EE66 pKa = 4.83YY67 pKa = 11.18NVSIYY72 pKa = 10.65KK73 pKa = 10.08DD74 pKa = 3.16AYY76 pKa = 9.39NNYY79 pKa = 9.65HH80 pKa = 5.54VFSSEE85 pKa = 3.49GHH87 pKa = 6.75DD88 pKa = 3.32IITNFSEE95 pKa = 4.61AEE97 pKa = 3.63GDD99 pKa = 3.6KK100 pKa = 10.61IYY102 pKa = 11.31VLDD105 pKa = 4.74PQPADD110 pKa = 4.0PINRR114 pKa = 11.84IKK116 pKa = 10.72PIDD119 pKa = 4.3PIIYY123 pKa = 9.82RR124 pKa = 11.84GSADD128 pKa = 3.75NIKK131 pKa = 9.73TILGANTSQYY141 pKa = 9.88TLSNSSLMLEE151 pKa = 4.36SNILQNPLYY160 pKa = 10.93NNLSNLGADD169 pKa = 3.37TSGFGLL175 pKa = 4.02
Molecular weight: 19.04 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1D2P4A1|A0A1D2P4A1_9CYAN Transmembrane sensor domain-containing protein (Fragment) OS=Scytonema millei VB511283 OX=1245923 GN=QH73_015475 PE=4 SV=1
MM1 pKa = 7.34RR2 pKa = 11.84TFIIIWIGQLVSLLGSGLTGFALSIWVYY30 pKa = 11.23KK31 pKa = 8.84NTNSVTQFALISVLTMLPNILISPLAGGLGDD62 pKa = 3.28RR63 pKa = 11.84RR64 pKa = 11.84NRR66 pKa = 11.84RR67 pKa = 11.84KK68 pKa = 10.35LIILSNCGSGLSILSLALLLFIGHH92 pKa = 6.51LEE94 pKa = 3.42IWHH97 pKa = 6.66IYY99 pKa = 8.73IATAASSAFHH109 pKa = 6.97ALQGPAYY116 pKa = 9.88DD117 pKa = 4.33AVITQLIPPQHH128 pKa = 6.37LVRR131 pKa = 11.84ANGMFQLGLAVAQLISPVLGGILLVTIQIQGIFILDD167 pKa = 4.86GITFIFCLVMLLLVRR182 pKa = 11.84LPKK185 pKa = 10.45PITTTKK191 pKa = 11.11AEE193 pKa = 4.21VPKK196 pKa = 10.86NLLWHH201 pKa = 6.58DD202 pKa = 3.97TLVGWNYY209 pKa = 8.02ITTRR213 pKa = 11.84RR214 pKa = 11.84GLFWLLILNCVYY226 pKa = 10.77HH227 pKa = 6.09FVEE230 pKa = 5.86GIAVVLSTPLFLSFTSTAVLGTMLSIGGSGMILGTALISTWGGAKK275 pKa = 9.46RR276 pKa = 11.84RR277 pKa = 11.84IYY279 pKa = 10.96NVFGFMLLGGLCMLVAGLRR298 pKa = 11.84RR299 pKa = 11.84NVLLCTVAIFLHH311 pKa = 6.53FFSLPIVVSSRR322 pKa = 11.84QAIFQSKK329 pKa = 8.76IAADD333 pKa = 3.24VRR335 pKa = 11.84ARR337 pKa = 11.84VFAVNRR343 pKa = 11.84MLINLSLLLAYY354 pKa = 10.31LVAGPLADD362 pKa = 4.33RR363 pKa = 11.84VFEE366 pKa = 4.22PLLTINGPLAGSVGKK381 pKa = 10.36IIGVGPGRR389 pKa = 11.84GIGLLFIIMGMSLIVAIVTSYY410 pKa = 11.18LSPRR414 pKa = 11.84LRR416 pKa = 11.84LVEE419 pKa = 4.25SEE421 pKa = 4.2LPNVISHH428 pKa = 7.04
MM1 pKa = 7.34RR2 pKa = 11.84TFIIIWIGQLVSLLGSGLTGFALSIWVYY30 pKa = 11.23KK31 pKa = 8.84NTNSVTQFALISVLTMLPNILISPLAGGLGDD62 pKa = 3.28RR63 pKa = 11.84RR64 pKa = 11.84NRR66 pKa = 11.84RR67 pKa = 11.84KK68 pKa = 10.35LIILSNCGSGLSILSLALLLFIGHH92 pKa = 6.51LEE94 pKa = 3.42IWHH97 pKa = 6.66IYY99 pKa = 8.73IATAASSAFHH109 pKa = 6.97ALQGPAYY116 pKa = 9.88DD117 pKa = 4.33AVITQLIPPQHH128 pKa = 6.37LVRR131 pKa = 11.84ANGMFQLGLAVAQLISPVLGGILLVTIQIQGIFILDD167 pKa = 4.86GITFIFCLVMLLLVRR182 pKa = 11.84LPKK185 pKa = 10.45PITTTKK191 pKa = 11.11AEE193 pKa = 4.21VPKK196 pKa = 10.86NLLWHH201 pKa = 6.58DD202 pKa = 3.97TLVGWNYY209 pKa = 8.02ITTRR213 pKa = 11.84RR214 pKa = 11.84GLFWLLILNCVYY226 pKa = 10.77HH227 pKa = 6.09FVEE230 pKa = 5.86GIAVVLSTPLFLSFTSTAVLGTMLSIGGSGMILGTALISTWGGAKK275 pKa = 9.46RR276 pKa = 11.84RR277 pKa = 11.84IYY279 pKa = 10.96NVFGFMLLGGLCMLVAGLRR298 pKa = 11.84RR299 pKa = 11.84NVLLCTVAIFLHH311 pKa = 6.53FFSLPIVVSSRR322 pKa = 11.84QAIFQSKK329 pKa = 8.76IAADD333 pKa = 3.24VRR335 pKa = 11.84ARR337 pKa = 11.84VFAVNRR343 pKa = 11.84MLINLSLLLAYY354 pKa = 10.31LVAGPLADD362 pKa = 4.33RR363 pKa = 11.84VFEE366 pKa = 4.22PLLTINGPLAGSVGKK381 pKa = 10.36IIGVGPGRR389 pKa = 11.84GIGLLFIIMGMSLIVAIVTSYY410 pKa = 11.18LSPRR414 pKa = 11.84LRR416 pKa = 11.84LVEE419 pKa = 4.25SEE421 pKa = 4.2LPNVISHH428 pKa = 7.04
Molecular weight: 46.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1457382 |
30 |
2762 |
294.4 |
32.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.892 ± 0.04 | 1.035 ± 0.012 |
4.73 ± 0.027 | 6.347 ± 0.038 |
4.085 ± 0.023 | 6.46 ± 0.037 |
1.89 ± 0.018 | 6.532 ± 0.026 |
5.191 ± 0.031 | 10.94 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.817 ± 0.014 | 4.412 ± 0.028 |
4.588 ± 0.024 | 5.413 ± 0.032 |
5.185 ± 0.029 | 6.458 ± 0.031 |
5.625 ± 0.025 | 6.838 ± 0.027 |
1.428 ± 0.016 | 3.132 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
