
Betaproteobacteria bacterium SCGC AG-212-J23
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; unclassified Betaproteobacteria
Average proteome isoelectric point is 7.45
Get precalculated fractions of proteins
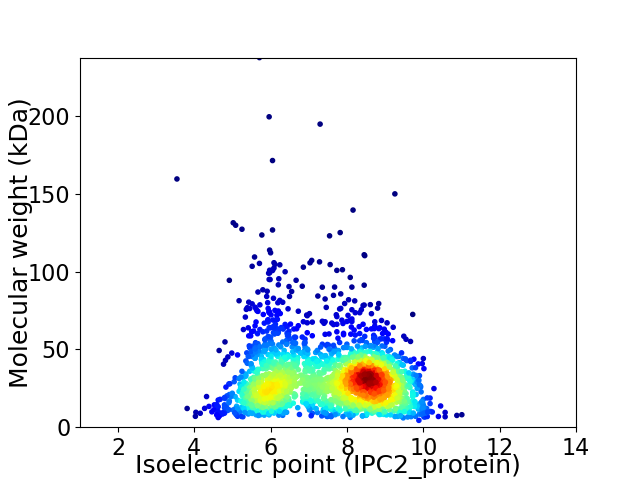
Virtual 2D-PAGE plot for 2067 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A177R0S4|A0A177R0S4_9PROT Enoyl-CoA hydratase OS=Betaproteobacteria bacterium SCGC AG-212-J23 OX=1799662 GN=AYO46_11035 PE=3 SV=1
MM1 pKa = 7.66TEE3 pKa = 3.78NQVQYY8 pKa = 8.74KK9 pKa = 8.09TWMCLICGFVYY20 pKa = 10.69DD21 pKa = 4.88EE22 pKa = 4.62AAGLPDD28 pKa = 3.87EE29 pKa = 5.79GIAPGTRR36 pKa = 11.84WDD38 pKa = 4.21DD39 pKa = 3.52VPMNWTCPEE48 pKa = 3.86CGARR52 pKa = 11.84KK53 pKa = 9.51EE54 pKa = 3.97DD55 pKa = 3.81FEE57 pKa = 4.37MVEE60 pKa = 4.0II61 pKa = 4.86
MM1 pKa = 7.66TEE3 pKa = 3.78NQVQYY8 pKa = 8.74KK9 pKa = 8.09TWMCLICGFVYY20 pKa = 10.69DD21 pKa = 4.88EE22 pKa = 4.62AAGLPDD28 pKa = 3.87EE29 pKa = 5.79GIAPGTRR36 pKa = 11.84WDD38 pKa = 4.21DD39 pKa = 3.52VPMNWTCPEE48 pKa = 3.86CGARR52 pKa = 11.84KK53 pKa = 9.51EE54 pKa = 3.97DD55 pKa = 3.81FEE57 pKa = 4.37MVEE60 pKa = 4.0II61 pKa = 4.86
Molecular weight: 6.99 kDa
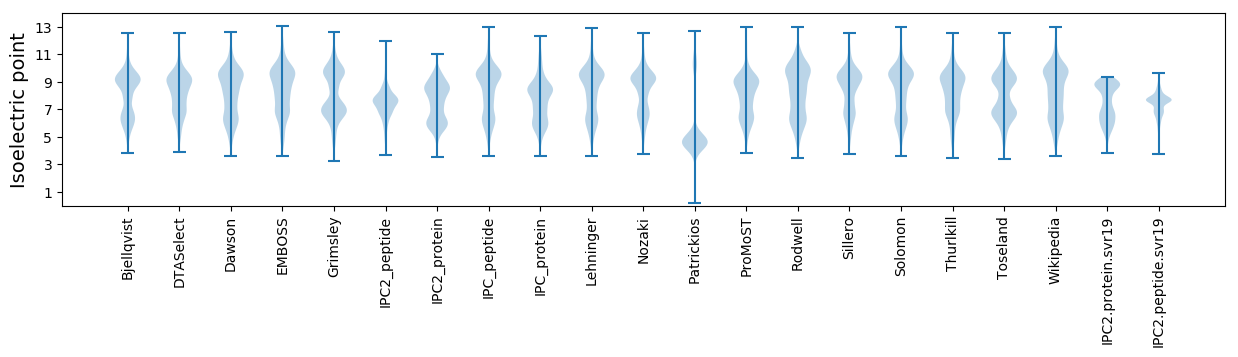
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A177QU73|A0A177QU73_9PROT CMD domain-containing protein (Fragment) OS=Betaproteobacteria bacterium SCGC AG-212-J23 OX=1799662 GN=AYO46_10620 PE=4 SV=1
MM1 pKa = 7.41SFAEE5 pKa = 5.72KK6 pKa = 10.18LVAWQRR12 pKa = 11.84RR13 pKa = 11.84QGRR16 pKa = 11.84HH17 pKa = 5.54DD18 pKa = 4.92LPWQGTRR25 pKa = 11.84DD26 pKa = 3.83PYY28 pKa = 10.85RR29 pKa = 11.84IWLSEE34 pKa = 3.93VMLQQTQVATVIPYY48 pKa = 8.3YY49 pKa = 10.73EE50 pKa = 4.53RR51 pKa = 11.84FLKK54 pKa = 10.24KK55 pKa = 10.08YY56 pKa = 9.9PSVEE60 pKa = 4.06FLASASQEE68 pKa = 3.88EE69 pKa = 4.68VLQLWSGLGYY79 pKa = 9.3YY80 pKa = 10.52ARR82 pKa = 11.84GRR84 pKa = 11.84NLHH87 pKa = 6.31AAAKK91 pKa = 9.25EE92 pKa = 3.64IARR95 pKa = 11.84NGFPKK100 pKa = 10.46DD101 pKa = 3.1IAKK104 pKa = 10.49LPGVGRR110 pKa = 11.84STAAAIAVFAFGEE123 pKa = 4.21RR124 pKa = 11.84AAIMDD129 pKa = 4.19GNAKK133 pKa = 9.74RR134 pKa = 11.84VLARR138 pKa = 11.84VFGVPDD144 pKa = 4.6PDD146 pKa = 3.06WDD148 pKa = 3.75LAEE151 pKa = 5.21RR152 pKa = 11.84EE153 pKa = 4.4LPQKK157 pKa = 10.66DD158 pKa = 3.5VAVYY162 pKa = 8.13TQALMDD168 pKa = 4.99LGATVCTRR176 pKa = 11.84RR177 pKa = 11.84PDD179 pKa = 3.55CARR182 pKa = 11.84CPVQRR187 pKa = 11.84GCVARR192 pKa = 11.84RR193 pKa = 11.84KK194 pKa = 10.4DD195 pKa = 3.56RR196 pKa = 11.84VAEE199 pKa = 4.11LPAPRR204 pKa = 11.84VRR206 pKa = 11.84KK207 pKa = 9.45VLPLRR212 pKa = 11.84HH213 pKa = 5.82ATWSILKK220 pKa = 10.56RR221 pKa = 11.84NDD223 pKa = 3.12QILLEE228 pKa = 4.4RR229 pKa = 11.84RR230 pKa = 11.84PSSGIWGGLWCFPEE244 pKa = 4.07AAKK247 pKa = 10.59PGGRR251 pKa = 11.84KK252 pKa = 9.13LPPIEE257 pKa = 5.19HH258 pKa = 6.82GFTHH262 pKa = 6.88FRR264 pKa = 11.84LKK266 pKa = 10.03IQPVLRR272 pKa = 11.84EE273 pKa = 3.7VRR275 pKa = 11.84KK276 pKa = 10.09RR277 pKa = 11.84IPAGPAQRR285 pKa = 11.84WMRR288 pKa = 11.84LADD291 pKa = 3.5VASAAVPTPVRR302 pKa = 11.84RR303 pKa = 11.84LISSLRR309 pKa = 3.54
MM1 pKa = 7.41SFAEE5 pKa = 5.72KK6 pKa = 10.18LVAWQRR12 pKa = 11.84RR13 pKa = 11.84QGRR16 pKa = 11.84HH17 pKa = 5.54DD18 pKa = 4.92LPWQGTRR25 pKa = 11.84DD26 pKa = 3.83PYY28 pKa = 10.85RR29 pKa = 11.84IWLSEE34 pKa = 3.93VMLQQTQVATVIPYY48 pKa = 8.3YY49 pKa = 10.73EE50 pKa = 4.53RR51 pKa = 11.84FLKK54 pKa = 10.24KK55 pKa = 10.08YY56 pKa = 9.9PSVEE60 pKa = 4.06FLASASQEE68 pKa = 3.88EE69 pKa = 4.68VLQLWSGLGYY79 pKa = 9.3YY80 pKa = 10.52ARR82 pKa = 11.84GRR84 pKa = 11.84NLHH87 pKa = 6.31AAAKK91 pKa = 9.25EE92 pKa = 3.64IARR95 pKa = 11.84NGFPKK100 pKa = 10.46DD101 pKa = 3.1IAKK104 pKa = 10.49LPGVGRR110 pKa = 11.84STAAAIAVFAFGEE123 pKa = 4.21RR124 pKa = 11.84AAIMDD129 pKa = 4.19GNAKK133 pKa = 9.74RR134 pKa = 11.84VLARR138 pKa = 11.84VFGVPDD144 pKa = 4.6PDD146 pKa = 3.06WDD148 pKa = 3.75LAEE151 pKa = 5.21RR152 pKa = 11.84EE153 pKa = 4.4LPQKK157 pKa = 10.66DD158 pKa = 3.5VAVYY162 pKa = 8.13TQALMDD168 pKa = 4.99LGATVCTRR176 pKa = 11.84RR177 pKa = 11.84PDD179 pKa = 3.55CARR182 pKa = 11.84CPVQRR187 pKa = 11.84GCVARR192 pKa = 11.84RR193 pKa = 11.84KK194 pKa = 10.4DD195 pKa = 3.56RR196 pKa = 11.84VAEE199 pKa = 4.11LPAPRR204 pKa = 11.84VRR206 pKa = 11.84KK207 pKa = 9.45VLPLRR212 pKa = 11.84HH213 pKa = 5.82ATWSILKK220 pKa = 10.56RR221 pKa = 11.84NDD223 pKa = 3.12QILLEE228 pKa = 4.4RR229 pKa = 11.84RR230 pKa = 11.84PSSGIWGGLWCFPEE244 pKa = 4.07AAKK247 pKa = 10.59PGGRR251 pKa = 11.84KK252 pKa = 9.13LPPIEE257 pKa = 5.19HH258 pKa = 6.82GFTHH262 pKa = 6.88FRR264 pKa = 11.84LKK266 pKa = 10.03IQPVLRR272 pKa = 11.84EE273 pKa = 3.7VRR275 pKa = 11.84KK276 pKa = 10.09RR277 pKa = 11.84IPAGPAQRR285 pKa = 11.84WMRR288 pKa = 11.84LADD291 pKa = 3.5VASAAVPTPVRR302 pKa = 11.84RR303 pKa = 11.84LISSLRR309 pKa = 3.54
Molecular weight: 34.93 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
629598 |
37 |
2126 |
304.6 |
33.29 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.175 ± 0.071 | 0.908 ± 0.018 |
5.001 ± 0.03 | 5.885 ± 0.051 |
3.846 ± 0.035 | 8.341 ± 0.052 |
2.04 ± 0.026 | 4.689 ± 0.037 |
4.442 ± 0.051 | 10.374 ± 0.06 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.402 ± 0.028 | 2.597 ± 0.033 |
5.158 ± 0.036 | 3.248 ± 0.032 |
7.201 ± 0.06 | 5.213 ± 0.04 |
4.793 ± 0.059 | 7.768 ± 0.041 |
1.414 ± 0.024 | 2.504 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
