
Changuinola virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Sedoreovirinae; Orbivirus
Average proteome isoelectric point is 6.49
Get precalculated fractions of proteins
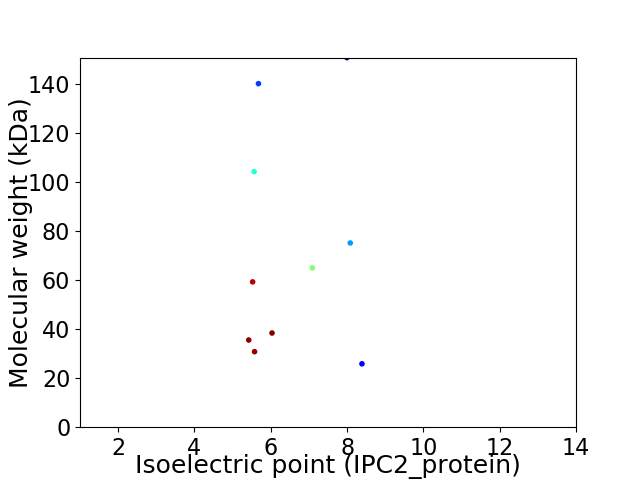
Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
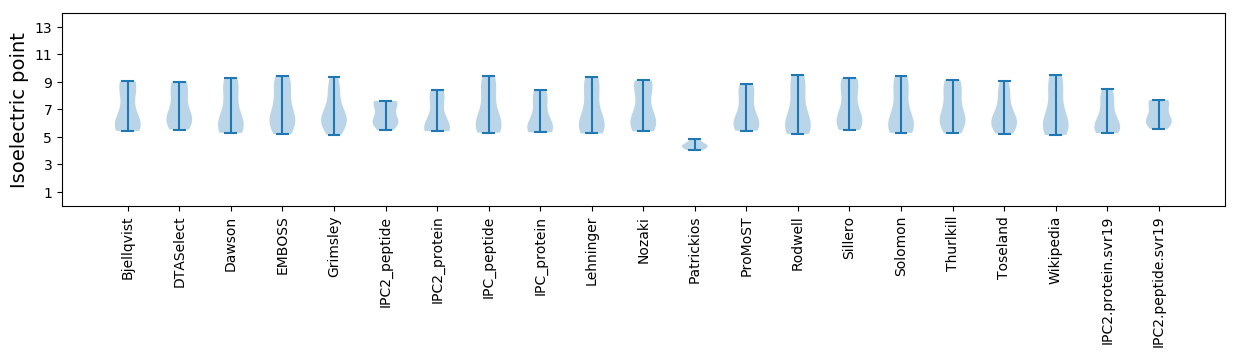
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|U5YL69|U5YL69_9REOV Core protein VP7 OS=Changuinola virus OX=40052 GN=CGLV_VP7 PE=3 SV=1
MM1 pKa = 7.4EE2 pKa = 5.5SKK4 pKa = 10.37PKK6 pKa = 10.28RR7 pKa = 11.84VFTKK11 pKa = 8.18TVCVLDD17 pKa = 4.6AYY19 pKa = 10.83GKK21 pKa = 6.63TFCGNVAKK29 pKa = 10.63LCNKK33 pKa = 8.62PYY35 pKa = 10.66CLIKK39 pKa = 9.96IGRR42 pKa = 11.84VVSIEE47 pKa = 4.18AVDD50 pKa = 3.65NPRR53 pKa = 11.84PKK55 pKa = 10.57SYY57 pKa = 10.44ILQIEE62 pKa = 4.22QVGSYY67 pKa = 10.0RR68 pKa = 11.84IQDD71 pKa = 3.65GQDD74 pKa = 3.43NISLIITNQGVEE86 pKa = 4.13ATVEE90 pKa = 4.02RR91 pKa = 11.84WEE93 pKa = 3.91EE94 pKa = 3.75WKK96 pKa = 10.86FEE98 pKa = 4.13NLSPIPMATMIMVADD113 pKa = 3.74KK114 pKa = 10.98RR115 pKa = 11.84VDD117 pKa = 3.43AEE119 pKa = 4.47VKK121 pKa = 9.58FVKK124 pKa = 10.48GMGIIAPYY132 pKa = 8.86TKK134 pKa = 10.77NEE136 pKa = 3.67MDD138 pKa = 3.65RR139 pKa = 11.84RR140 pKa = 11.84DD141 pKa = 3.76VPDD144 pKa = 4.18LPGVMPSSMGVKK156 pKa = 9.74EE157 pKa = 4.11LRR159 pKa = 11.84EE160 pKa = 3.69KK161 pKa = 10.81LRR163 pKa = 11.84NEE165 pKa = 3.89RR166 pKa = 11.84EE167 pKa = 3.78QGKK170 pKa = 9.27VRR172 pKa = 11.84EE173 pKa = 4.27SEE175 pKa = 3.93IVRR178 pKa = 11.84PAVVNHH184 pKa = 5.65VVEE187 pKa = 4.23QPKK190 pKa = 9.28WEE192 pKa = 4.48DD193 pKa = 3.52LDD195 pKa = 3.92TKK197 pKa = 10.52IAAVTDD203 pKa = 3.82WNADD207 pKa = 3.47VPSEE211 pKa = 4.13IPHH214 pKa = 6.21GSSEE218 pKa = 4.14EE219 pKa = 3.99EE220 pKa = 4.0EE221 pKa = 4.56EE222 pKa = 4.31EE223 pKa = 4.17GGEE226 pKa = 4.17YY227 pKa = 9.74LTGKK231 pKa = 9.69YY232 pKa = 7.73MKK234 pKa = 10.04KK235 pKa = 10.24VSTIVKK241 pKa = 10.05NPDD244 pKa = 3.21PSVLGMAFGYY254 pKa = 9.22PKK256 pKa = 10.32TSGPFDD262 pKa = 4.79RR263 pKa = 11.84IIVEE267 pKa = 5.15KK268 pKa = 9.9EE269 pKa = 4.11CLWTGVPIYY278 pKa = 10.87DD279 pKa = 4.01VDD281 pKa = 3.96QTDD284 pKa = 2.62KK285 pKa = 11.12SYY287 pKa = 10.65RR288 pKa = 11.84LVMIGEE294 pKa = 4.07ATKK297 pKa = 11.08YY298 pKa = 10.19LLAVKK303 pKa = 9.27GQNYY307 pKa = 8.55FVLPAGAGSAA317 pKa = 3.72
MM1 pKa = 7.4EE2 pKa = 5.5SKK4 pKa = 10.37PKK6 pKa = 10.28RR7 pKa = 11.84VFTKK11 pKa = 8.18TVCVLDD17 pKa = 4.6AYY19 pKa = 10.83GKK21 pKa = 6.63TFCGNVAKK29 pKa = 10.63LCNKK33 pKa = 8.62PYY35 pKa = 10.66CLIKK39 pKa = 9.96IGRR42 pKa = 11.84VVSIEE47 pKa = 4.18AVDD50 pKa = 3.65NPRR53 pKa = 11.84PKK55 pKa = 10.57SYY57 pKa = 10.44ILQIEE62 pKa = 4.22QVGSYY67 pKa = 10.0RR68 pKa = 11.84IQDD71 pKa = 3.65GQDD74 pKa = 3.43NISLIITNQGVEE86 pKa = 4.13ATVEE90 pKa = 4.02RR91 pKa = 11.84WEE93 pKa = 3.91EE94 pKa = 3.75WKK96 pKa = 10.86FEE98 pKa = 4.13NLSPIPMATMIMVADD113 pKa = 3.74KK114 pKa = 10.98RR115 pKa = 11.84VDD117 pKa = 3.43AEE119 pKa = 4.47VKK121 pKa = 9.58FVKK124 pKa = 10.48GMGIIAPYY132 pKa = 8.86TKK134 pKa = 10.77NEE136 pKa = 3.67MDD138 pKa = 3.65RR139 pKa = 11.84RR140 pKa = 11.84DD141 pKa = 3.76VPDD144 pKa = 4.18LPGVMPSSMGVKK156 pKa = 9.74EE157 pKa = 4.11LRR159 pKa = 11.84EE160 pKa = 3.69KK161 pKa = 10.81LRR163 pKa = 11.84NEE165 pKa = 3.89RR166 pKa = 11.84EE167 pKa = 3.78QGKK170 pKa = 9.27VRR172 pKa = 11.84EE173 pKa = 4.27SEE175 pKa = 3.93IVRR178 pKa = 11.84PAVVNHH184 pKa = 5.65VVEE187 pKa = 4.23QPKK190 pKa = 9.28WEE192 pKa = 4.48DD193 pKa = 3.52LDD195 pKa = 3.92TKK197 pKa = 10.52IAAVTDD203 pKa = 3.82WNADD207 pKa = 3.47VPSEE211 pKa = 4.13IPHH214 pKa = 6.21GSSEE218 pKa = 4.14EE219 pKa = 3.99EE220 pKa = 4.0EE221 pKa = 4.56EE222 pKa = 4.31EE223 pKa = 4.17GGEE226 pKa = 4.17YY227 pKa = 9.74LTGKK231 pKa = 9.69YY232 pKa = 7.73MKK234 pKa = 10.04KK235 pKa = 10.24VSTIVKK241 pKa = 10.05NPDD244 pKa = 3.21PSVLGMAFGYY254 pKa = 9.22PKK256 pKa = 10.32TSGPFDD262 pKa = 4.79RR263 pKa = 11.84IIVEE267 pKa = 5.15KK268 pKa = 9.9EE269 pKa = 4.11CLWTGVPIYY278 pKa = 10.87DD279 pKa = 4.01VDD281 pKa = 3.96QTDD284 pKa = 2.62KK285 pKa = 11.12SYY287 pKa = 10.65RR288 pKa = 11.84LVMIGEE294 pKa = 4.07ATKK297 pKa = 11.08YY298 pKa = 10.19LLAVKK303 pKa = 9.27GQNYY307 pKa = 8.55FVLPAGAGSAA317 pKa = 3.72
Molecular weight: 35.51 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|U5NU19|U5NU19_9REOV Outer capsid protein VP2 OS=Changuinola virus OX=40052 GN=CGLV_VP2 PE=3 SV=1
MM1 pKa = 7.56LSEE4 pKa = 3.89LAARR8 pKa = 11.84FEE10 pKa = 4.36AGKK13 pKa = 9.84QRR15 pKa = 11.84EE16 pKa = 4.23LEE18 pKa = 4.15EE19 pKa = 4.41VNMLNEE25 pKa = 4.26TAVVPYY31 pKa = 8.85TRR33 pKa = 11.84PPSYY37 pKa = 11.08APTAPTTFAPAHH49 pKa = 6.18LSLNILNNAMSNGTAKK65 pKa = 10.05TNALKK70 pKa = 10.5EE71 pKa = 4.18EE72 pKa = 4.19KK73 pKa = 10.36VAFGSYY79 pKa = 10.82AEE81 pKa = 4.35AFRR84 pKa = 11.84DD85 pKa = 4.09EE86 pKa = 4.72PAVQQIKK93 pKa = 9.82SHH95 pKa = 5.52VNEE98 pKa = 4.05QIIPKK103 pKa = 10.06LKK105 pKa = 10.55RR106 pKa = 11.84EE107 pKa = 3.69LAGYY111 pKa = 8.88KK112 pKa = 9.4KK113 pKa = 10.45KK114 pKa = 10.45RR115 pKa = 11.84WLVHH119 pKa = 4.87LTMLIAAGVALFTSLGTLVKK139 pKa = 10.19DD140 pKa = 3.53VQIRR144 pKa = 11.84IPGNNSTAGANEE156 pKa = 4.57YY157 pKa = 10.17IHH159 pKa = 7.01LPTWYY164 pKa = 8.69TSLGAIFGIVNLGATGLMISCARR187 pKa = 11.84IEE189 pKa = 3.96KK190 pKa = 10.62SYY192 pKa = 11.43DD193 pKa = 3.08EE194 pKa = 5.34SIAFLKK200 pKa = 10.48KK201 pKa = 10.15EE202 pKa = 3.94LMKK205 pKa = 10.69KK206 pKa = 10.44RR207 pKa = 11.84SYY209 pKa = 11.42NDD211 pKa = 4.13AIRR214 pKa = 11.84MSIKK218 pKa = 10.89DD219 pKa = 3.62MGSLTNLFSEE229 pKa = 4.89EE230 pKa = 3.8QHH232 pKa = 5.35QQ233 pKa = 3.54
MM1 pKa = 7.56LSEE4 pKa = 3.89LAARR8 pKa = 11.84FEE10 pKa = 4.36AGKK13 pKa = 9.84QRR15 pKa = 11.84EE16 pKa = 4.23LEE18 pKa = 4.15EE19 pKa = 4.41VNMLNEE25 pKa = 4.26TAVVPYY31 pKa = 8.85TRR33 pKa = 11.84PPSYY37 pKa = 11.08APTAPTTFAPAHH49 pKa = 6.18LSLNILNNAMSNGTAKK65 pKa = 10.05TNALKK70 pKa = 10.5EE71 pKa = 4.18EE72 pKa = 4.19KK73 pKa = 10.36VAFGSYY79 pKa = 10.82AEE81 pKa = 4.35AFRR84 pKa = 11.84DD85 pKa = 4.09EE86 pKa = 4.72PAVQQIKK93 pKa = 9.82SHH95 pKa = 5.52VNEE98 pKa = 4.05QIIPKK103 pKa = 10.06LKK105 pKa = 10.55RR106 pKa = 11.84EE107 pKa = 3.69LAGYY111 pKa = 8.88KK112 pKa = 9.4KK113 pKa = 10.45KK114 pKa = 10.45RR115 pKa = 11.84WLVHH119 pKa = 4.87LTMLIAAGVALFTSLGTLVKK139 pKa = 10.19DD140 pKa = 3.53VQIRR144 pKa = 11.84IPGNNSTAGANEE156 pKa = 4.57YY157 pKa = 10.17IHH159 pKa = 7.01LPTWYY164 pKa = 8.69TSLGAIFGIVNLGATGLMISCARR187 pKa = 11.84IEE189 pKa = 3.96KK190 pKa = 10.62SYY192 pKa = 11.43DD193 pKa = 3.08EE194 pKa = 5.34SIAFLKK200 pKa = 10.48KK201 pKa = 10.15EE202 pKa = 3.94LMKK205 pKa = 10.69KK206 pKa = 10.44RR207 pKa = 11.84SYY209 pKa = 11.42NDD211 pKa = 4.13AIRR214 pKa = 11.84MSIKK218 pKa = 10.89DD219 pKa = 3.62MGSLTNLFSEE229 pKa = 4.89EE230 pKa = 3.8QHH232 pKa = 5.35QQ233 pKa = 3.54
Molecular weight: 25.82 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6321 |
233 |
1309 |
632.1 |
72.43 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.138 ± 0.66 | 1.155 ± 0.281 |
5.759 ± 0.332 | 7.293 ± 0.512 |
4.018 ± 0.354 | 5.885 ± 0.396 |
2.421 ± 0.233 | 7.087 ± 0.326 |
6.186 ± 0.707 | 8.448 ± 0.321 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.654 ± 0.399 | 4.888 ± 0.271 |
3.734 ± 0.347 | 3.623 ± 0.277 |
6.423 ± 0.496 | 5.474 ± 0.492 |
5.363 ± 0.398 | 7.182 ± 0.326 |
1.202 ± 0.181 | 4.066 ± 0.344 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
