
Barbel circovirus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; Circovirus
Average proteome isoelectric point is 8.98
Get precalculated fractions of proteins
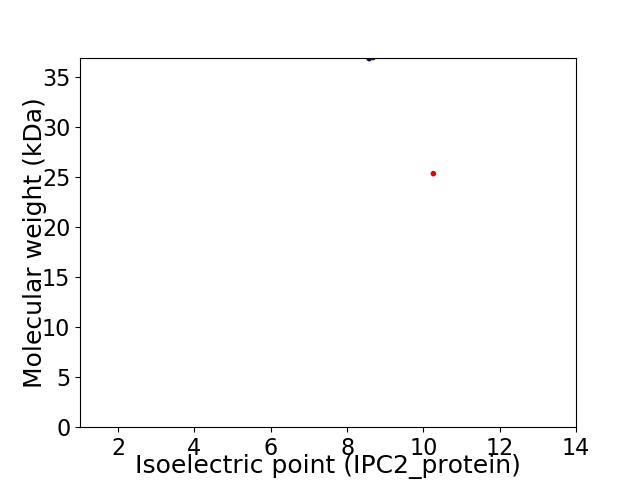
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F5BSB5|F5BSB5_9CIRC Capsid protein OS=Barbel circovirus OX=759938 GN=cap PE=3 SV=1
MM1 pKa = 7.75PKK3 pKa = 9.01ATSKK7 pKa = 9.18TRR9 pKa = 11.84HH10 pKa = 5.44LQNPRR15 pKa = 11.84QDD17 pKa = 3.35GPRR20 pKa = 11.84RR21 pKa = 11.84EE22 pKa = 4.15QPVKK26 pKa = 10.25RR27 pKa = 11.84WCFTLNNPTAEE38 pKa = 4.32EE39 pKa = 4.01RR40 pKa = 11.84RR41 pKa = 11.84HH42 pKa = 4.71IQQIITADD50 pKa = 3.52SVDD53 pKa = 4.0FAVIGNEE60 pKa = 3.95IGDD63 pKa = 3.75SGTPHH68 pKa = 6.49LQGFLNMKK76 pKa = 7.28TKK78 pKa = 10.51RR79 pKa = 11.84RR80 pKa = 11.84LGTMKK85 pKa = 10.1KK86 pKa = 8.63WFNARR91 pKa = 11.84AQYY94 pKa = 9.66EE95 pKa = 4.27AAKK98 pKa = 9.29GTDD101 pKa = 3.62LQNDD105 pKa = 4.57EE106 pKa = 4.7YY107 pKa = 9.78CTKK110 pKa = 10.96GGDD113 pKa = 3.06TYY115 pKa = 11.6LRR117 pKa = 11.84IGEE120 pKa = 4.24PGKK123 pKa = 10.12EE124 pKa = 3.56RR125 pKa = 11.84CRR127 pKa = 11.84NDD129 pKa = 3.11LQKK132 pKa = 11.12AIDD135 pKa = 3.72VVKK138 pKa = 10.5RR139 pKa = 11.84SSGSMRR145 pKa = 11.84AVAEE149 pKa = 4.09ACPATFIRR157 pKa = 11.84YY158 pKa = 8.59GRR160 pKa = 11.84GLRR163 pKa = 11.84DD164 pKa = 3.42YY165 pKa = 10.83ANVMQYY171 pKa = 10.48RR172 pKa = 11.84KK173 pKa = 9.61PRR175 pKa = 11.84DD176 pKa = 3.24FKK178 pKa = 11.06TEE180 pKa = 3.72VKK182 pKa = 10.61VYY184 pKa = 10.72VGDD187 pKa = 4.13PGCSKK192 pKa = 10.46SRR194 pKa = 11.84KK195 pKa = 9.41ASEE198 pKa = 4.2LCAGTTVYY206 pKa = 10.58YY207 pKa = 10.42KK208 pKa = 10.5PRR210 pKa = 11.84GMWWDD215 pKa = 4.0GYY217 pKa = 11.01DD218 pKa = 3.41GQEE221 pKa = 3.94NVIVDD226 pKa = 4.48DD227 pKa = 4.59FYY229 pKa = 11.78GWMPCDD235 pKa = 3.13EE236 pKa = 4.86LLRR239 pKa = 11.84VFDD242 pKa = 5.32RR243 pKa = 11.84YY244 pKa = 9.32PCKK247 pKa = 10.88VPVKK251 pKa = 8.87GAYY254 pKa = 9.61VEE256 pKa = 4.72FVSTAIYY263 pKa = 8.36VTSNKK268 pKa = 9.79HH269 pKa = 3.04VWQWYY274 pKa = 8.76KK275 pKa = 11.04FEE277 pKa = 4.81GFDD280 pKa = 3.3PAAVMRR286 pKa = 11.84RR287 pKa = 11.84VNVYY291 pKa = 10.17LVYY294 pKa = 10.96DD295 pKa = 3.64NAGEE299 pKa = 4.02RR300 pKa = 11.84FVNLRR305 pKa = 11.84EE306 pKa = 3.93SAMYY310 pKa = 10.6DD311 pKa = 3.67PVTMRR316 pKa = 11.84YY317 pKa = 9.75CYY319 pKa = 10.8
MM1 pKa = 7.75PKK3 pKa = 9.01ATSKK7 pKa = 9.18TRR9 pKa = 11.84HH10 pKa = 5.44LQNPRR15 pKa = 11.84QDD17 pKa = 3.35GPRR20 pKa = 11.84RR21 pKa = 11.84EE22 pKa = 4.15QPVKK26 pKa = 10.25RR27 pKa = 11.84WCFTLNNPTAEE38 pKa = 4.32EE39 pKa = 4.01RR40 pKa = 11.84RR41 pKa = 11.84HH42 pKa = 4.71IQQIITADD50 pKa = 3.52SVDD53 pKa = 4.0FAVIGNEE60 pKa = 3.95IGDD63 pKa = 3.75SGTPHH68 pKa = 6.49LQGFLNMKK76 pKa = 7.28TKK78 pKa = 10.51RR79 pKa = 11.84RR80 pKa = 11.84LGTMKK85 pKa = 10.1KK86 pKa = 8.63WFNARR91 pKa = 11.84AQYY94 pKa = 9.66EE95 pKa = 4.27AAKK98 pKa = 9.29GTDD101 pKa = 3.62LQNDD105 pKa = 4.57EE106 pKa = 4.7YY107 pKa = 9.78CTKK110 pKa = 10.96GGDD113 pKa = 3.06TYY115 pKa = 11.6LRR117 pKa = 11.84IGEE120 pKa = 4.24PGKK123 pKa = 10.12EE124 pKa = 3.56RR125 pKa = 11.84CRR127 pKa = 11.84NDD129 pKa = 3.11LQKK132 pKa = 11.12AIDD135 pKa = 3.72VVKK138 pKa = 10.5RR139 pKa = 11.84SSGSMRR145 pKa = 11.84AVAEE149 pKa = 4.09ACPATFIRR157 pKa = 11.84YY158 pKa = 8.59GRR160 pKa = 11.84GLRR163 pKa = 11.84DD164 pKa = 3.42YY165 pKa = 10.83ANVMQYY171 pKa = 10.48RR172 pKa = 11.84KK173 pKa = 9.61PRR175 pKa = 11.84DD176 pKa = 3.24FKK178 pKa = 11.06TEE180 pKa = 3.72VKK182 pKa = 10.61VYY184 pKa = 10.72VGDD187 pKa = 4.13PGCSKK192 pKa = 10.46SRR194 pKa = 11.84KK195 pKa = 9.41ASEE198 pKa = 4.2LCAGTTVYY206 pKa = 10.58YY207 pKa = 10.42KK208 pKa = 10.5PRR210 pKa = 11.84GMWWDD215 pKa = 4.0GYY217 pKa = 11.01DD218 pKa = 3.41GQEE221 pKa = 3.94NVIVDD226 pKa = 4.48DD227 pKa = 4.59FYY229 pKa = 11.78GWMPCDD235 pKa = 3.13EE236 pKa = 4.86LLRR239 pKa = 11.84VFDD242 pKa = 5.32RR243 pKa = 11.84YY244 pKa = 9.32PCKK247 pKa = 10.88VPVKK251 pKa = 8.87GAYY254 pKa = 9.61VEE256 pKa = 4.72FVSTAIYY263 pKa = 8.36VTSNKK268 pKa = 9.79HH269 pKa = 3.04VWQWYY274 pKa = 8.76KK275 pKa = 11.04FEE277 pKa = 4.81GFDD280 pKa = 3.3PAAVMRR286 pKa = 11.84RR287 pKa = 11.84VNVYY291 pKa = 10.17LVYY294 pKa = 10.96DD295 pKa = 3.64NAGEE299 pKa = 4.02RR300 pKa = 11.84FVNLRR305 pKa = 11.84EE306 pKa = 3.93SAMYY310 pKa = 10.6DD311 pKa = 3.67PVTMRR316 pKa = 11.84YY317 pKa = 9.75CYY319 pKa = 10.8
Molecular weight: 36.88 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F5BSB5|F5BSB5_9CIRC Capsid protein OS=Barbel circovirus OX=759938 GN=cap PE=3 SV=1
MM1 pKa = 7.59LLPRR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84ARR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84AGRR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84TRR19 pKa = 11.84GWRR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84KK25 pKa = 8.38YY26 pKa = 10.49VRR28 pKa = 11.84RR29 pKa = 11.84QRR31 pKa = 11.84HH32 pKa = 5.21RR33 pKa = 11.84GRR35 pKa = 11.84YY36 pKa = 8.33SYY38 pKa = 11.06FRR40 pKa = 11.84LRR42 pKa = 11.84AIKK45 pKa = 9.94HH46 pKa = 6.29DD47 pKa = 3.7HH48 pKa = 5.8LTWNASNGMSSGIVFLMADD67 pKa = 3.43MSYY70 pKa = 10.67PKK72 pKa = 10.51NYY74 pKa = 8.12WDD76 pKa = 3.73YY77 pKa = 9.92YY78 pKa = 9.79KK79 pKa = 10.27ISKK82 pKa = 10.0VVVQLWPEE90 pKa = 4.15FNNVPINADD99 pKa = 3.65RR100 pKa = 11.84DD101 pKa = 3.74KK102 pKa = 11.47VLYY105 pKa = 10.65GSTAVDD111 pKa = 3.4YY112 pKa = 11.31DD113 pKa = 4.18DD114 pKa = 4.76ATMATSTADD123 pKa = 3.3PFKK126 pKa = 10.68DD127 pKa = 3.48YY128 pKa = 11.09SSRR131 pKa = 11.84KK132 pKa = 8.2MFISNRR138 pKa = 11.84THH140 pKa = 6.61IRR142 pKa = 11.84IFTPRR147 pKa = 11.84PNIEE151 pKa = 4.08IYY153 pKa = 10.44KK154 pKa = 10.47SSASTNYY161 pKa = 9.48GFLNIRR167 pKa = 11.84SGAWINTVHH176 pKa = 7.32DD177 pKa = 4.47GVPHH181 pKa = 6.13YY182 pKa = 10.42GLKK185 pKa = 9.62IWLPSNTGAPSAIGYY200 pKa = 9.61KK201 pKa = 10.42LIIKK205 pKa = 9.47YY206 pKa = 10.12YY207 pKa = 8.4VLFRR211 pKa = 11.84NRR213 pKa = 11.84II214 pKa = 3.49
MM1 pKa = 7.59LLPRR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84ARR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84AGRR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84TRR19 pKa = 11.84GWRR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84KK25 pKa = 8.38YY26 pKa = 10.49VRR28 pKa = 11.84RR29 pKa = 11.84QRR31 pKa = 11.84HH32 pKa = 5.21RR33 pKa = 11.84GRR35 pKa = 11.84YY36 pKa = 8.33SYY38 pKa = 11.06FRR40 pKa = 11.84LRR42 pKa = 11.84AIKK45 pKa = 9.94HH46 pKa = 6.29DD47 pKa = 3.7HH48 pKa = 5.8LTWNASNGMSSGIVFLMADD67 pKa = 3.43MSYY70 pKa = 10.67PKK72 pKa = 10.51NYY74 pKa = 8.12WDD76 pKa = 3.73YY77 pKa = 9.92YY78 pKa = 9.79KK79 pKa = 10.27ISKK82 pKa = 10.0VVVQLWPEE90 pKa = 4.15FNNVPINADD99 pKa = 3.65RR100 pKa = 11.84DD101 pKa = 3.74KK102 pKa = 11.47VLYY105 pKa = 10.65GSTAVDD111 pKa = 3.4YY112 pKa = 11.31DD113 pKa = 4.18DD114 pKa = 4.76ATMATSTADD123 pKa = 3.3PFKK126 pKa = 10.68DD127 pKa = 3.48YY128 pKa = 11.09SSRR131 pKa = 11.84KK132 pKa = 8.2MFISNRR138 pKa = 11.84THH140 pKa = 6.61IRR142 pKa = 11.84IFTPRR147 pKa = 11.84PNIEE151 pKa = 4.08IYY153 pKa = 10.44KK154 pKa = 10.47SSASTNYY161 pKa = 9.48GFLNIRR167 pKa = 11.84SGAWINTVHH176 pKa = 7.32DD177 pKa = 4.47GVPHH181 pKa = 6.13YY182 pKa = 10.42GLKK185 pKa = 9.62IWLPSNTGAPSAIGYY200 pKa = 9.61KK201 pKa = 10.42LIIKK205 pKa = 9.47YY206 pKa = 10.12YY207 pKa = 8.4VLFRR211 pKa = 11.84NRR213 pKa = 11.84II214 pKa = 3.49
Molecular weight: 25.37 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
533 |
214 |
319 |
266.5 |
31.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.754 ± 0.135 | 1.689 ± 1.075 |
6.004 ± 0.55 | 3.565 ± 1.675 |
3.752 ± 0.009 | 6.754 ± 0.73 |
1.876 ± 0.591 | 4.878 ± 1.655 |
6.379 ± 0.491 | 4.878 ± 0.465 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.002 ± 0.126 | 5.066 ± 0.643 |
4.878 ± 0.131 | 2.627 ± 1.078 |
10.882 ± 1.7 | 5.441 ± 1.594 |
5.441 ± 0.192 | 6.942 ± 1.148 |
2.439 ± 0.232 | 6.754 ± 0.46 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
