
Flavobacterium phage FPSV-S1
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Myoviridae; unclassified Myoviridae
Average proteome isoelectric point is 6.65
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 43 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
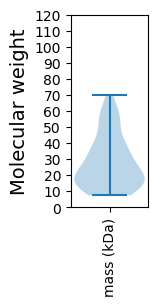
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A6M2VB84|A0A6M2VB84_9CAUD Uncharacterized protein OS=Flavobacterium phage FPSV-S1 OX=2575345 PE=4 SV=1
MM1 pKa = 7.58AKK3 pKa = 10.15KK4 pKa = 10.4SIAILKK10 pKa = 9.69QYY12 pKa = 10.34YY13 pKa = 8.5LTGLRR18 pKa = 11.84PTQQNYY24 pKa = 10.24HH25 pKa = 6.76DD26 pKa = 4.17WLDD29 pKa = 3.87SFLHH33 pKa = 6.55KK34 pKa = 10.61DD35 pKa = 3.47DD36 pKa = 4.54GVLITSIVSDD46 pKa = 3.85PLIGNVTINYY56 pKa = 7.7TSGDD60 pKa = 3.61PFIFNVTPTSVPISAVTGLQDD81 pKa = 3.33VLNNFVLQQNGYY93 pKa = 10.09GLTQNNFTNEE103 pKa = 3.75DD104 pKa = 3.72VILLNEE110 pKa = 4.69LGTYY114 pKa = 10.44LDD116 pKa = 3.81YY117 pKa = 11.27LQNYY121 pKa = 9.24NGGLSVKK128 pKa = 10.63NIFIPSNLVLDD139 pKa = 4.02SAGADD144 pKa = 2.88IEE146 pKa = 4.92GFLSEE151 pKa = 4.83IEE153 pKa = 4.02PSFSINRR160 pKa = 11.84GQLLTVKK167 pKa = 10.1YY168 pKa = 10.46NRR170 pKa = 11.84AEE172 pKa = 4.05LSDD175 pKa = 3.78GSSALVHH182 pKa = 6.29CNAVFALPAGKK193 pKa = 10.38YY194 pKa = 9.93GFGFGDD200 pKa = 3.6VYY202 pKa = 11.18GSTVFDD208 pKa = 3.48SYY210 pKa = 11.28EE211 pKa = 3.86FKK213 pKa = 11.06LRR215 pKa = 11.84DD216 pKa = 3.53SAVVLEE222 pKa = 4.65DD223 pKa = 3.72EE224 pKa = 4.52YY225 pKa = 11.74SIFGLNAVDD234 pKa = 3.29IQGNLLTTSVQEE246 pKa = 4.29QFNQAAYY253 pKa = 10.07KK254 pKa = 10.33NITSIYY260 pKa = 8.9GKK262 pKa = 9.32PAALRR267 pKa = 11.84GRR269 pKa = 11.84DD270 pKa = 3.58FTNGYY275 pKa = 8.19SAQWNDD281 pKa = 3.7NTPMSWINDD290 pKa = 3.56EE291 pKa = 4.43IYY293 pKa = 10.54RR294 pKa = 11.84ISTNVNFSQLQDD306 pKa = 3.45GFSVICQIEE315 pKa = 4.24NQDD318 pKa = 4.54PITPSIMRR326 pKa = 11.84RR327 pKa = 11.84FYY329 pKa = 11.17LALNKK334 pKa = 10.4AQGSCVFSCWLEE346 pKa = 3.86EE347 pKa = 4.32VQSNTYY353 pKa = 10.19LKK355 pKa = 10.28RR356 pKa = 11.84KK357 pKa = 9.84KK358 pKa = 10.68FEE360 pKa = 4.01TDD362 pKa = 2.55ATNIPEE368 pKa = 4.36NFFKK372 pKa = 10.5IDD374 pKa = 3.35VEE376 pKa = 4.74VIINDD381 pKa = 4.21LAANNFNCLFNIDD394 pKa = 3.88GASFSLTQGNVTFTDD409 pKa = 3.73YY410 pKa = 11.36SGGYY414 pKa = 7.87GTTNYY419 pKa = 8.38PQNNRR424 pKa = 11.84ICWGNYY430 pKa = 7.89YY431 pKa = 9.29PAKK434 pKa = 10.16ILMSSFYY441 pKa = 10.4YY442 pKa = 10.01QINGIEE448 pKa = 4.4RR449 pKa = 11.84NFSLDD454 pKa = 3.27QPYY457 pKa = 10.27IPYY460 pKa = 8.81ATTDD464 pKa = 3.52GVFMNISPNNDD475 pKa = 3.31NFWDD479 pKa = 4.01LSKK482 pKa = 10.08TSSLYY487 pKa = 10.63VKK489 pKa = 10.04PSSDD493 pKa = 3.8VIRR496 pKa = 11.84ALDD499 pKa = 3.6FNDD502 pKa = 2.62IHH504 pKa = 7.37ALNNTEE510 pKa = 4.43VFDD513 pKa = 4.69LSMYY517 pKa = 9.37MSPNWRR523 pKa = 11.84GKK525 pKa = 10.66VFGQMNNGNLRR536 pKa = 11.84VFITGSYY543 pKa = 10.08KK544 pKa = 10.57GDD546 pKa = 3.32KK547 pKa = 9.82TDD549 pKa = 3.71LNTYY553 pKa = 10.06ILLDD557 pKa = 4.58KK558 pKa = 10.83IPEE561 pKa = 3.96FLFNRR566 pKa = 11.84DD567 pKa = 3.5YY568 pKa = 10.43QTSKK572 pKa = 10.92CLYY575 pKa = 8.32LQNIKK580 pKa = 10.63RR581 pKa = 11.84PTDD584 pKa = 3.73TILGAAVMGIEE595 pKa = 4.26NNLKK599 pKa = 10.12IKK601 pKa = 10.67VNNFDD606 pKa = 3.62INGFEE611 pKa = 4.15IKK613 pKa = 8.69EE614 pKa = 4.3TLVTFDD620 pKa = 4.83LLNN623 pKa = 3.79
MM1 pKa = 7.58AKK3 pKa = 10.15KK4 pKa = 10.4SIAILKK10 pKa = 9.69QYY12 pKa = 10.34YY13 pKa = 8.5LTGLRR18 pKa = 11.84PTQQNYY24 pKa = 10.24HH25 pKa = 6.76DD26 pKa = 4.17WLDD29 pKa = 3.87SFLHH33 pKa = 6.55KK34 pKa = 10.61DD35 pKa = 3.47DD36 pKa = 4.54GVLITSIVSDD46 pKa = 3.85PLIGNVTINYY56 pKa = 7.7TSGDD60 pKa = 3.61PFIFNVTPTSVPISAVTGLQDD81 pKa = 3.33VLNNFVLQQNGYY93 pKa = 10.09GLTQNNFTNEE103 pKa = 3.75DD104 pKa = 3.72VILLNEE110 pKa = 4.69LGTYY114 pKa = 10.44LDD116 pKa = 3.81YY117 pKa = 11.27LQNYY121 pKa = 9.24NGGLSVKK128 pKa = 10.63NIFIPSNLVLDD139 pKa = 4.02SAGADD144 pKa = 2.88IEE146 pKa = 4.92GFLSEE151 pKa = 4.83IEE153 pKa = 4.02PSFSINRR160 pKa = 11.84GQLLTVKK167 pKa = 10.1YY168 pKa = 10.46NRR170 pKa = 11.84AEE172 pKa = 4.05LSDD175 pKa = 3.78GSSALVHH182 pKa = 6.29CNAVFALPAGKK193 pKa = 10.38YY194 pKa = 9.93GFGFGDD200 pKa = 3.6VYY202 pKa = 11.18GSTVFDD208 pKa = 3.48SYY210 pKa = 11.28EE211 pKa = 3.86FKK213 pKa = 11.06LRR215 pKa = 11.84DD216 pKa = 3.53SAVVLEE222 pKa = 4.65DD223 pKa = 3.72EE224 pKa = 4.52YY225 pKa = 11.74SIFGLNAVDD234 pKa = 3.29IQGNLLTTSVQEE246 pKa = 4.29QFNQAAYY253 pKa = 10.07KK254 pKa = 10.33NITSIYY260 pKa = 8.9GKK262 pKa = 9.32PAALRR267 pKa = 11.84GRR269 pKa = 11.84DD270 pKa = 3.58FTNGYY275 pKa = 8.19SAQWNDD281 pKa = 3.7NTPMSWINDD290 pKa = 3.56EE291 pKa = 4.43IYY293 pKa = 10.54RR294 pKa = 11.84ISTNVNFSQLQDD306 pKa = 3.45GFSVICQIEE315 pKa = 4.24NQDD318 pKa = 4.54PITPSIMRR326 pKa = 11.84RR327 pKa = 11.84FYY329 pKa = 11.17LALNKK334 pKa = 10.4AQGSCVFSCWLEE346 pKa = 3.86EE347 pKa = 4.32VQSNTYY353 pKa = 10.19LKK355 pKa = 10.28RR356 pKa = 11.84KK357 pKa = 9.84KK358 pKa = 10.68FEE360 pKa = 4.01TDD362 pKa = 2.55ATNIPEE368 pKa = 4.36NFFKK372 pKa = 10.5IDD374 pKa = 3.35VEE376 pKa = 4.74VIINDD381 pKa = 4.21LAANNFNCLFNIDD394 pKa = 3.88GASFSLTQGNVTFTDD409 pKa = 3.73YY410 pKa = 11.36SGGYY414 pKa = 7.87GTTNYY419 pKa = 8.38PQNNRR424 pKa = 11.84ICWGNYY430 pKa = 7.89YY431 pKa = 9.29PAKK434 pKa = 10.16ILMSSFYY441 pKa = 10.4YY442 pKa = 10.01QINGIEE448 pKa = 4.4RR449 pKa = 11.84NFSLDD454 pKa = 3.27QPYY457 pKa = 10.27IPYY460 pKa = 8.81ATTDD464 pKa = 3.52GVFMNISPNNDD475 pKa = 3.31NFWDD479 pKa = 4.01LSKK482 pKa = 10.08TSSLYY487 pKa = 10.63VKK489 pKa = 10.04PSSDD493 pKa = 3.8VIRR496 pKa = 11.84ALDD499 pKa = 3.6FNDD502 pKa = 2.62IHH504 pKa = 7.37ALNNTEE510 pKa = 4.43VFDD513 pKa = 4.69LSMYY517 pKa = 9.37MSPNWRR523 pKa = 11.84GKK525 pKa = 10.66VFGQMNNGNLRR536 pKa = 11.84VFITGSYY543 pKa = 10.08KK544 pKa = 10.57GDD546 pKa = 3.32KK547 pKa = 9.82TDD549 pKa = 3.71LNTYY553 pKa = 10.06ILLDD557 pKa = 4.58KK558 pKa = 10.83IPEE561 pKa = 3.96FLFNRR566 pKa = 11.84DD567 pKa = 3.5YY568 pKa = 10.43QTSKK572 pKa = 10.92CLYY575 pKa = 8.32LQNIKK580 pKa = 10.63RR581 pKa = 11.84PTDD584 pKa = 3.73TILGAAVMGIEE595 pKa = 4.26NNLKK599 pKa = 10.12IKK601 pKa = 10.67VNNFDD606 pKa = 3.62INGFEE611 pKa = 4.15IKK613 pKa = 8.69EE614 pKa = 4.3TLVTFDD620 pKa = 4.83LLNN623 pKa = 3.79
Molecular weight: 70.18 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6M2VB90|A0A6M2VB90_9CAUD Uncharacterized protein OS=Flavobacterium phage FPSV-S1 OX=2575345 PE=4 SV=1
MM1 pKa = 6.93MSFRR5 pKa = 11.84FKK7 pKa = 11.03EE8 pKa = 3.82KK9 pKa = 9.92IAAFKK14 pKa = 10.73AMKK17 pKa = 8.62RR18 pKa = 11.84TLPIVVANAAKK29 pKa = 10.3NHH31 pKa = 5.72FLEE34 pKa = 5.01GFRR37 pKa = 11.84KK38 pKa = 10.33GGFTDD43 pKa = 3.91EE44 pKa = 6.05SFDD47 pKa = 3.33PWVKK51 pKa = 10.55RR52 pKa = 11.84KK53 pKa = 10.09AKK55 pKa = 10.08AKK57 pKa = 9.77RR58 pKa = 11.84NAGRR62 pKa = 11.84ALLVYY67 pKa = 9.88KK68 pKa = 10.45GHH70 pKa = 6.92LRR72 pKa = 11.84RR73 pKa = 11.84SIRR76 pKa = 11.84VISASFNKK84 pKa = 9.93IEE86 pKa = 4.2VGSTGTKK93 pKa = 8.52YY94 pKa = 10.82ASRR97 pKa = 11.84HH98 pKa = 4.31NQGLDD103 pKa = 3.36GMPKK107 pKa = 9.46RR108 pKa = 11.84QFIGKK113 pKa = 9.45SGQLHH118 pKa = 5.93KK119 pKa = 10.49KK120 pKa = 8.52ISKK123 pKa = 10.12IIQNKK128 pKa = 9.58IKK130 pKa = 10.69DD131 pKa = 3.7SLLL134 pKa = 3.7
MM1 pKa = 6.93MSFRR5 pKa = 11.84FKK7 pKa = 11.03EE8 pKa = 3.82KK9 pKa = 9.92IAAFKK14 pKa = 10.73AMKK17 pKa = 8.62RR18 pKa = 11.84TLPIVVANAAKK29 pKa = 10.3NHH31 pKa = 5.72FLEE34 pKa = 5.01GFRR37 pKa = 11.84KK38 pKa = 10.33GGFTDD43 pKa = 3.91EE44 pKa = 6.05SFDD47 pKa = 3.33PWVKK51 pKa = 10.55RR52 pKa = 11.84KK53 pKa = 10.09AKK55 pKa = 10.08AKK57 pKa = 9.77RR58 pKa = 11.84NAGRR62 pKa = 11.84ALLVYY67 pKa = 9.88KK68 pKa = 10.45GHH70 pKa = 6.92LRR72 pKa = 11.84RR73 pKa = 11.84SIRR76 pKa = 11.84VISASFNKK84 pKa = 9.93IEE86 pKa = 4.2VGSTGTKK93 pKa = 8.52YY94 pKa = 10.82ASRR97 pKa = 11.84HH98 pKa = 4.31NQGLDD103 pKa = 3.36GMPKK107 pKa = 9.46RR108 pKa = 11.84QFIGKK113 pKa = 9.45SGQLHH118 pKa = 5.93KK119 pKa = 10.49KK120 pKa = 8.52ISKK123 pKa = 10.12IIQNKK128 pKa = 9.58IKK130 pKa = 10.69DD131 pKa = 3.7SLLL134 pKa = 3.7
Molecular weight: 15.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
10714 |
64 |
623 |
249.2 |
28.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.842 ± 0.561 | 0.831 ± 0.116 |
6.291 ± 0.267 | 6.683 ± 0.403 |
5.124 ± 0.317 | 6.225 ± 0.35 |
0.989 ± 0.101 | 8.27 ± 0.271 |
9.091 ± 0.563 | 8.83 ± 0.331 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.605 ± 0.156 | 6.478 ± 0.398 |
2.893 ± 0.169 | 3.64 ± 0.128 |
3.509 ± 0.213 | 5.628 ± 0.237 |
5.852 ± 0.226 | 6.412 ± 0.214 |
1.139 ± 0.105 | 3.668 ± 0.294 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
