
Thermococcus kodakarensis (strain ATCC BAA-918 / JCM 12380 / KOD1) (Pyrococcus kodakaraensis (strain KOD1))
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Thermococci; Thermococcales; Thermococcaceae; Thermococcus; Thermococcus kodakarensis
Average proteome isoelectric point is 6.54
Get precalculated fractions of proteins
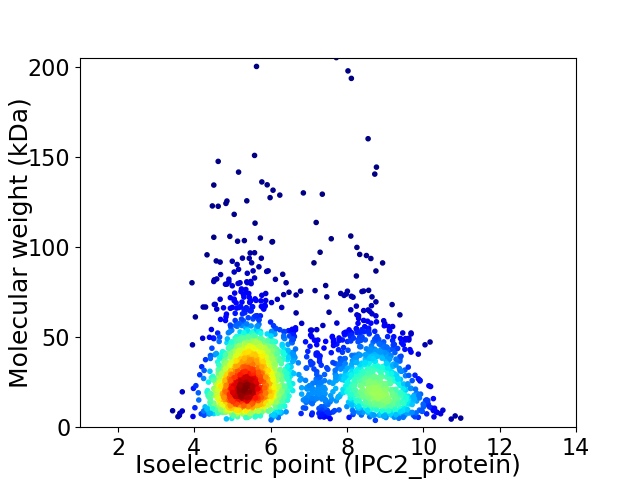
Virtual 2D-PAGE plot for 2301 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q5JHL8|PSB2_THEKO Proteasome subunit beta 2 OS=Thermococcus kodakarensis (strain ATCC BAA-918 / JCM 12380 / KOD1) OX=69014 GN=psmB2 PE=3 SV=1
MM1 pKa = 7.62SKK3 pKa = 9.19FTGLLLILILLGAAYY18 pKa = 9.31IYY20 pKa = 7.64YY21 pKa = 8.47TNPHH25 pKa = 6.83IIEE28 pKa = 5.5DD29 pKa = 3.87LTKK32 pKa = 10.45MKK34 pKa = 10.81DD35 pKa = 3.24SFPNGFDD42 pKa = 3.21SGGTTTHH49 pKa = 6.52SSGGSGFSEE58 pKa = 4.23TSSGNHH64 pKa = 5.44SEE66 pKa = 4.19SFPGEE71 pKa = 4.25KK72 pKa = 10.22GWVDD76 pKa = 3.21SDD78 pKa = 3.37NDD80 pKa = 3.5GRR82 pKa = 11.84KK83 pKa = 9.5DD84 pKa = 3.54VYY86 pKa = 10.9IGDD89 pKa = 4.83DD90 pKa = 3.62YY91 pKa = 12.23AEE93 pKa = 4.22VPTTNGSVLFVDD105 pKa = 4.19RR106 pKa = 11.84NKK108 pKa = 10.83DD109 pKa = 3.38GKK111 pKa = 10.4IDD113 pKa = 5.02AIYY116 pKa = 10.87LDD118 pKa = 3.83TTGDD122 pKa = 3.43GSYY125 pKa = 9.6DD126 pKa = 3.31TAYY129 pKa = 11.05LDD131 pKa = 3.51EE132 pKa = 5.91DD133 pKa = 4.81YY134 pKa = 11.37NGKK137 pKa = 8.12TDD139 pKa = 3.1TWRR142 pKa = 11.84TTFNGVDD149 pKa = 3.36SYY151 pKa = 12.18AWDD154 pKa = 3.36ITGDD158 pKa = 4.56GIPDD162 pKa = 3.59VYY164 pKa = 11.21DD165 pKa = 3.55SNGDD169 pKa = 3.69GKK171 pKa = 11.13VDD173 pKa = 3.23AWDD176 pKa = 3.86TNSDD180 pKa = 3.97GIIDD184 pKa = 4.24EE185 pKa = 4.78RR186 pKa = 11.84DD187 pKa = 2.87VDD189 pKa = 4.15YY190 pKa = 11.51DD191 pKa = 3.76GTPDD195 pKa = 3.23LHH197 pKa = 8.36DD198 pKa = 3.83YY199 pKa = 11.48DD200 pKa = 4.44FDD202 pKa = 4.2GVFDD206 pKa = 3.72EE207 pKa = 5.16FEE209 pKa = 4.97RR210 pKa = 11.84NVTLYY215 pKa = 10.23PPEE218 pKa = 4.4SANGTSGAFLCPDD231 pKa = 3.99NKK233 pKa = 10.86DD234 pKa = 3.01DD235 pKa = 4.38AYY237 pKa = 11.41RR238 pKa = 11.84LFVQAYY244 pKa = 9.11NNVTSLQSSGASDD257 pKa = 4.21EE258 pKa = 4.93EE259 pKa = 4.1IQEE262 pKa = 4.82AYY264 pKa = 10.35AKK266 pKa = 8.71YY267 pKa = 9.87QKK269 pKa = 10.83ARR271 pKa = 11.84ACYY274 pKa = 9.64EE275 pKa = 3.98SFLTSTTSAASSSGMRR291 pKa = 11.84EE292 pKa = 3.8IKK294 pKa = 9.63TVTLDD299 pKa = 3.46TEE301 pKa = 4.27GTAGVKK307 pKa = 10.58FSTGEE312 pKa = 3.66LKK314 pKa = 10.61GYY316 pKa = 10.44NEE318 pKa = 3.93ISDD321 pKa = 4.34NVWEE325 pKa = 4.73DD326 pKa = 2.91VDD328 pKa = 5.49LVAEE332 pKa = 4.78PWCVEE337 pKa = 3.81YY338 pKa = 9.39PALLGHH344 pKa = 7.24WIDD347 pKa = 4.2LGEE350 pKa = 4.56GNLDD354 pKa = 3.48TLDD357 pKa = 3.86PSEE360 pKa = 5.16IPTSGYY366 pKa = 6.73PTGEE370 pKa = 3.63IAEE373 pKa = 4.55EE374 pKa = 3.67IKK376 pKa = 10.28MGHH379 pKa = 6.1VYY381 pKa = 10.92VNLNSDD387 pKa = 3.59GTLTAFEE394 pKa = 4.63LVSHH398 pKa = 6.96EE399 pKa = 4.12KK400 pKa = 10.13TGDD403 pKa = 3.29CSHH406 pKa = 7.53RR407 pKa = 11.84ITIRR411 pKa = 11.84YY412 pKa = 9.04SNLGGSS418 pKa = 3.77
MM1 pKa = 7.62SKK3 pKa = 9.19FTGLLLILILLGAAYY18 pKa = 9.31IYY20 pKa = 7.64YY21 pKa = 8.47TNPHH25 pKa = 6.83IIEE28 pKa = 5.5DD29 pKa = 3.87LTKK32 pKa = 10.45MKK34 pKa = 10.81DD35 pKa = 3.24SFPNGFDD42 pKa = 3.21SGGTTTHH49 pKa = 6.52SSGGSGFSEE58 pKa = 4.23TSSGNHH64 pKa = 5.44SEE66 pKa = 4.19SFPGEE71 pKa = 4.25KK72 pKa = 10.22GWVDD76 pKa = 3.21SDD78 pKa = 3.37NDD80 pKa = 3.5GRR82 pKa = 11.84KK83 pKa = 9.5DD84 pKa = 3.54VYY86 pKa = 10.9IGDD89 pKa = 4.83DD90 pKa = 3.62YY91 pKa = 12.23AEE93 pKa = 4.22VPTTNGSVLFVDD105 pKa = 4.19RR106 pKa = 11.84NKK108 pKa = 10.83DD109 pKa = 3.38GKK111 pKa = 10.4IDD113 pKa = 5.02AIYY116 pKa = 10.87LDD118 pKa = 3.83TTGDD122 pKa = 3.43GSYY125 pKa = 9.6DD126 pKa = 3.31TAYY129 pKa = 11.05LDD131 pKa = 3.51EE132 pKa = 5.91DD133 pKa = 4.81YY134 pKa = 11.37NGKK137 pKa = 8.12TDD139 pKa = 3.1TWRR142 pKa = 11.84TTFNGVDD149 pKa = 3.36SYY151 pKa = 12.18AWDD154 pKa = 3.36ITGDD158 pKa = 4.56GIPDD162 pKa = 3.59VYY164 pKa = 11.21DD165 pKa = 3.55SNGDD169 pKa = 3.69GKK171 pKa = 11.13VDD173 pKa = 3.23AWDD176 pKa = 3.86TNSDD180 pKa = 3.97GIIDD184 pKa = 4.24EE185 pKa = 4.78RR186 pKa = 11.84DD187 pKa = 2.87VDD189 pKa = 4.15YY190 pKa = 11.51DD191 pKa = 3.76GTPDD195 pKa = 3.23LHH197 pKa = 8.36DD198 pKa = 3.83YY199 pKa = 11.48DD200 pKa = 4.44FDD202 pKa = 4.2GVFDD206 pKa = 3.72EE207 pKa = 5.16FEE209 pKa = 4.97RR210 pKa = 11.84NVTLYY215 pKa = 10.23PPEE218 pKa = 4.4SANGTSGAFLCPDD231 pKa = 3.99NKK233 pKa = 10.86DD234 pKa = 3.01DD235 pKa = 4.38AYY237 pKa = 11.41RR238 pKa = 11.84LFVQAYY244 pKa = 9.11NNVTSLQSSGASDD257 pKa = 4.21EE258 pKa = 4.93EE259 pKa = 4.1IQEE262 pKa = 4.82AYY264 pKa = 10.35AKK266 pKa = 8.71YY267 pKa = 9.87QKK269 pKa = 10.83ARR271 pKa = 11.84ACYY274 pKa = 9.64EE275 pKa = 3.98SFLTSTTSAASSSGMRR291 pKa = 11.84EE292 pKa = 3.8IKK294 pKa = 9.63TVTLDD299 pKa = 3.46TEE301 pKa = 4.27GTAGVKK307 pKa = 10.58FSTGEE312 pKa = 3.66LKK314 pKa = 10.61GYY316 pKa = 10.44NEE318 pKa = 3.93ISDD321 pKa = 4.34NVWEE325 pKa = 4.73DD326 pKa = 2.91VDD328 pKa = 5.49LVAEE332 pKa = 4.78PWCVEE337 pKa = 3.81YY338 pKa = 9.39PALLGHH344 pKa = 7.24WIDD347 pKa = 4.2LGEE350 pKa = 4.56GNLDD354 pKa = 3.48TLDD357 pKa = 3.86PSEE360 pKa = 5.16IPTSGYY366 pKa = 6.73PTGEE370 pKa = 3.63IAEE373 pKa = 4.55EE374 pKa = 3.67IKK376 pKa = 10.28MGHH379 pKa = 6.1VYY381 pKa = 10.92VNLNSDD387 pKa = 3.59GTLTAFEE394 pKa = 4.63LVSHH398 pKa = 6.96EE399 pKa = 4.12KK400 pKa = 10.13TGDD403 pKa = 3.29CSHH406 pKa = 7.53RR407 pKa = 11.84ITIRR411 pKa = 11.84YY412 pKa = 9.04SNLGGSS418 pKa = 3.77
Molecular weight: 45.66 kDa
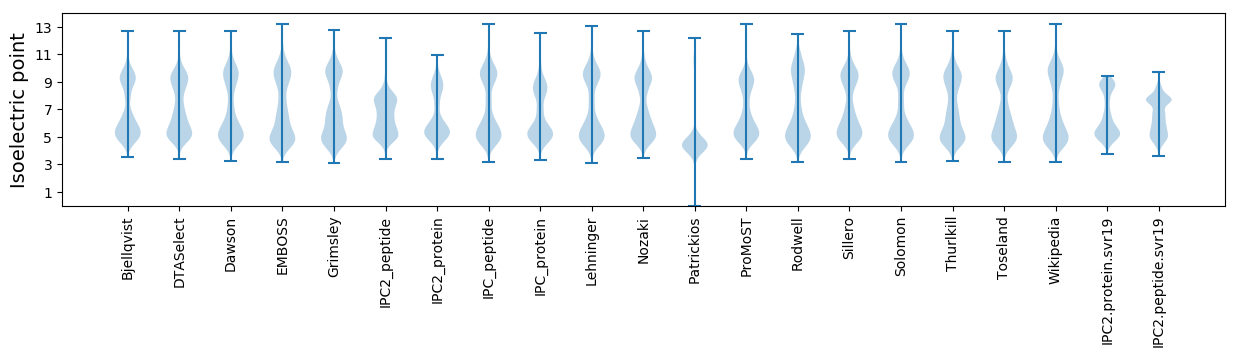
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q5JEZ6|Q5JEZ6_THEKO Non-specific serine/threonine protein kinase OS=Thermococcus kodakarensis (strain ATCC BAA-918 / JCM 12380 / KOD1) OX=69014 GN=TK0679 PE=3 SV=1
MM1 pKa = 7.48KK2 pKa = 10.23RR3 pKa = 11.84RR4 pKa = 11.84PRR6 pKa = 11.84KK7 pKa = 7.59WKK9 pKa = 10.02KK10 pKa = 8.58KK11 pKa = 10.27GRR13 pKa = 11.84MRR15 pKa = 11.84WKK17 pKa = 9.51WIKK20 pKa = 10.46KK21 pKa = 8.96RR22 pKa = 11.84IRR24 pKa = 11.84RR25 pKa = 11.84LKK27 pKa = 8.25RR28 pKa = 11.84QRR30 pKa = 11.84RR31 pKa = 11.84KK32 pKa = 10.18EE33 pKa = 3.73RR34 pKa = 11.84GLII37 pKa = 3.73
MM1 pKa = 7.48KK2 pKa = 10.23RR3 pKa = 11.84RR4 pKa = 11.84PRR6 pKa = 11.84KK7 pKa = 7.59WKK9 pKa = 10.02KK10 pKa = 8.58KK11 pKa = 10.27GRR13 pKa = 11.84MRR15 pKa = 11.84WKK17 pKa = 9.51WIKK20 pKa = 10.46KK21 pKa = 8.96RR22 pKa = 11.84IRR24 pKa = 11.84RR25 pKa = 11.84LKK27 pKa = 8.25RR28 pKa = 11.84QRR30 pKa = 11.84RR31 pKa = 11.84KK32 pKa = 10.18EE33 pKa = 3.73RR34 pKa = 11.84GLII37 pKa = 3.73
Molecular weight: 5.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
637680 |
33 |
1798 |
277.1 |
31.19 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.419 ± 0.052 | 0.531 ± 0.016 |
4.733 ± 0.041 | 8.826 ± 0.078 |
4.33 ± 0.039 | 7.584 ± 0.052 |
1.592 ± 0.019 | 6.961 ± 0.049 |
6.868 ± 0.055 | 10.472 ± 0.071 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.338 ± 0.023 | 3.214 ± 0.034 |
4.388 ± 0.03 | 1.823 ± 0.027 |
5.84 ± 0.05 | 5.047 ± 0.043 |
4.615 ± 0.042 | 8.292 ± 0.043 |
1.296 ± 0.02 | 3.832 ± 0.035 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
