
Eubacterium brachy ATCC 33089
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Eubacteriales incertae sedis; Eubacteriales Family XIII. Incertae Sedis; [Eubacterium] brachy
Average proteome isoelectric point is 6.83
Get precalculated fractions of proteins
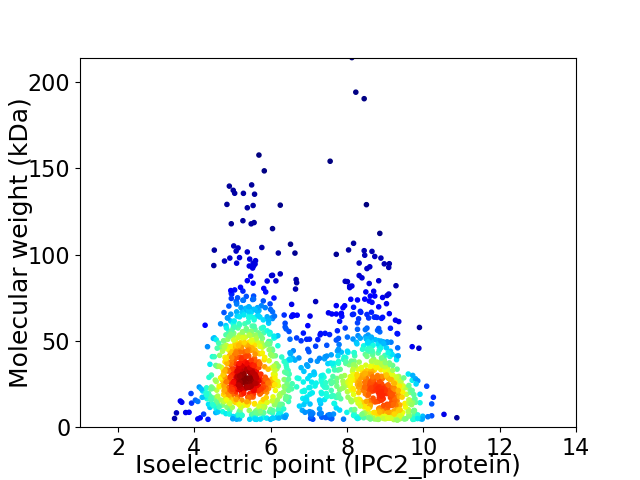
Virtual 2D-PAGE plot for 1479 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
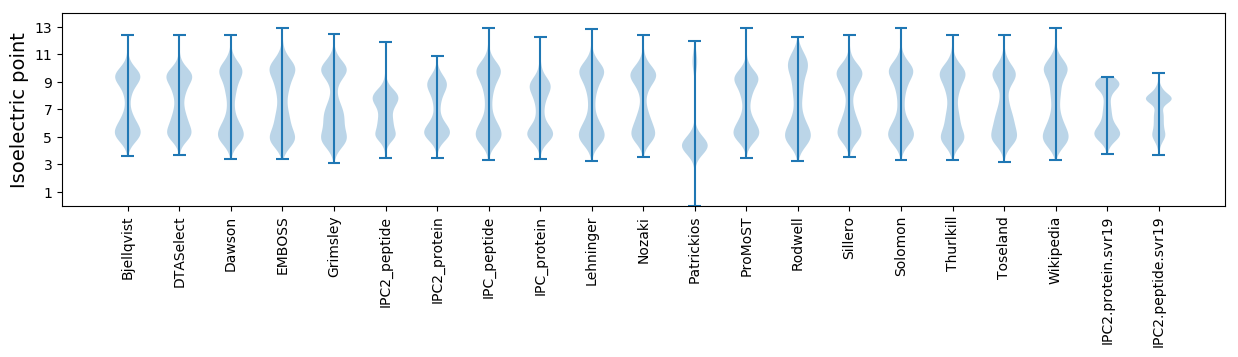
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|V1CNI1|V1CNI1_9FIRM NAD-dependent protein deacetylase OS=Eubacterium brachy ATCC 33089 OX=1321814 GN=cobB PE=3 SV=1
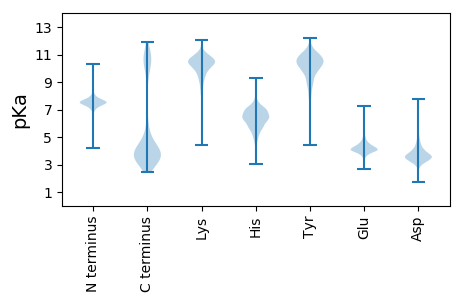
MM1 pKa = 7.34FFDD4 pKa = 4.48EE5 pKa = 5.68DD6 pKa = 5.2DD7 pKa = 4.1PVSWSVDD14 pKa = 2.71IGFSEE19 pKa = 4.67EE20 pKa = 4.21DD21 pKa = 3.52FGDD24 pKa = 3.74EE25 pKa = 4.12TVSPVICINPIDD37 pKa = 3.76TDD39 pKa = 3.89KK40 pKa = 11.75NSVEE44 pKa = 4.07EE45 pKa = 3.93LVGEE49 pKa = 4.2KK50 pKa = 10.52FKK52 pKa = 11.35VATVEE57 pKa = 4.03EE58 pKa = 4.6CADD61 pKa = 4.21RR62 pKa = 11.84EE63 pKa = 4.39DD64 pKa = 3.14TFYY67 pKa = 10.43IYY69 pKa = 10.4EE70 pKa = 4.22FEE72 pKa = 4.27PMVSYY77 pKa = 10.19EE78 pKa = 4.14VEE80 pKa = 3.89VLEE83 pKa = 5.25IKK85 pKa = 10.74DD86 pKa = 3.33SMAHH90 pKa = 5.04VRR92 pKa = 11.84CSGIMIVDD100 pKa = 5.34GYY102 pKa = 10.8SDD104 pKa = 4.41PYY106 pKa = 9.4VQEE109 pKa = 4.02TFEE112 pKa = 4.56IDD114 pKa = 2.63SWIPVIEE121 pKa = 5.11SVDD124 pKa = 3.25DD125 pKa = 3.54WAKK128 pKa = 11.0FEE130 pKa = 4.24NCC132 pKa = 4.35
MM1 pKa = 7.34FFDD4 pKa = 4.48EE5 pKa = 5.68DD6 pKa = 5.2DD7 pKa = 4.1PVSWSVDD14 pKa = 2.71IGFSEE19 pKa = 4.67EE20 pKa = 4.21DD21 pKa = 3.52FGDD24 pKa = 3.74EE25 pKa = 4.12TVSPVICINPIDD37 pKa = 3.76TDD39 pKa = 3.89KK40 pKa = 11.75NSVEE44 pKa = 4.07EE45 pKa = 3.93LVGEE49 pKa = 4.2KK50 pKa = 10.52FKK52 pKa = 11.35VATVEE57 pKa = 4.03EE58 pKa = 4.6CADD61 pKa = 4.21RR62 pKa = 11.84EE63 pKa = 4.39DD64 pKa = 3.14TFYY67 pKa = 10.43IYY69 pKa = 10.4EE70 pKa = 4.22FEE72 pKa = 4.27PMVSYY77 pKa = 10.19EE78 pKa = 4.14VEE80 pKa = 3.89VLEE83 pKa = 5.25IKK85 pKa = 10.74DD86 pKa = 3.33SMAHH90 pKa = 5.04VRR92 pKa = 11.84CSGIMIVDD100 pKa = 5.34GYY102 pKa = 10.8SDD104 pKa = 4.41PYY106 pKa = 9.4VQEE109 pKa = 4.02TFEE112 pKa = 4.56IDD114 pKa = 2.63SWIPVIEE121 pKa = 5.11SVDD124 pKa = 3.25DD125 pKa = 3.54WAKK128 pKa = 11.0FEE130 pKa = 4.24NCC132 pKa = 4.35
Molecular weight: 15.18 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|V1CKN3|V1CKN3_9FIRM tRNA-specific adenosine deaminase OS=Eubacterium brachy ATCC 33089 OX=1321814 GN=tadA PE=3 SV=1
MM1 pKa = 7.73LIGVVAFFAKK11 pKa = 10.49DD12 pKa = 3.34IIYY15 pKa = 10.45GSMVFIIVCLLSFLIGQRR33 pKa = 11.84KK34 pKa = 7.24KK35 pKa = 8.45TIKK38 pKa = 10.63FIVFYY43 pKa = 10.6IFLLLFIVFSKK54 pKa = 10.16YY55 pKa = 10.35VPVTLSNMMLMIVLCMRR72 pKa = 11.84MCMPILLYY80 pKa = 10.74GQTFLRR86 pKa = 11.84TTPVSEE92 pKa = 4.32MVTGLYY98 pKa = 10.35AIRR101 pKa = 11.84IPRR104 pKa = 11.84SFIITFAVAMRR115 pKa = 11.84FFPTAKK121 pKa = 10.29EE122 pKa = 4.21EE123 pKa = 4.03ISHH126 pKa = 6.15VRR128 pKa = 11.84DD129 pKa = 3.27AMSLRR134 pKa = 11.84GIGFSWRR141 pKa = 11.84NLRR144 pKa = 11.84RR145 pKa = 11.84RR146 pKa = 11.84PGMVFEE152 pKa = 5.06GFITPLLVRR161 pKa = 11.84ASTIAEE167 pKa = 4.12EE168 pKa = 4.33LSAASITRR176 pKa = 11.84GLDD179 pKa = 3.04NPAPRR184 pKa = 11.84TAFIKK189 pKa = 10.92LNITVKK195 pKa = 10.11DD196 pKa = 3.54TFLTVIFTLLLISVVLLRR214 pKa = 11.84TIVGRR219 pKa = 11.84WW220 pKa = 2.99
MM1 pKa = 7.73LIGVVAFFAKK11 pKa = 10.49DD12 pKa = 3.34IIYY15 pKa = 10.45GSMVFIIVCLLSFLIGQRR33 pKa = 11.84KK34 pKa = 7.24KK35 pKa = 8.45TIKK38 pKa = 10.63FIVFYY43 pKa = 10.6IFLLLFIVFSKK54 pKa = 10.16YY55 pKa = 10.35VPVTLSNMMLMIVLCMRR72 pKa = 11.84MCMPILLYY80 pKa = 10.74GQTFLRR86 pKa = 11.84TTPVSEE92 pKa = 4.32MVTGLYY98 pKa = 10.35AIRR101 pKa = 11.84IPRR104 pKa = 11.84SFIITFAVAMRR115 pKa = 11.84FFPTAKK121 pKa = 10.29EE122 pKa = 4.21EE123 pKa = 4.03ISHH126 pKa = 6.15VRR128 pKa = 11.84DD129 pKa = 3.27AMSLRR134 pKa = 11.84GIGFSWRR141 pKa = 11.84NLRR144 pKa = 11.84RR145 pKa = 11.84RR146 pKa = 11.84PGMVFEE152 pKa = 5.06GFITPLLVRR161 pKa = 11.84ASTIAEE167 pKa = 4.12EE168 pKa = 4.33LSAASITRR176 pKa = 11.84GLDD179 pKa = 3.04NPAPRR184 pKa = 11.84TAFIKK189 pKa = 10.92LNITVKK195 pKa = 10.11DD196 pKa = 3.54TFLTVIFTLLLISVVLLRR214 pKa = 11.84TIVGRR219 pKa = 11.84WW220 pKa = 2.99
Molecular weight: 25.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
455806 |
36 |
1926 |
308.2 |
34.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.238 ± 0.069 | 1.196 ± 0.026 |
5.818 ± 0.056 | 7.593 ± 0.082 |
4.363 ± 0.05 | 6.998 ± 0.068 |
1.568 ± 0.025 | 8.928 ± 0.065 |
8.925 ± 0.07 | 8.409 ± 0.068 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.052 ± 0.032 | 5.204 ± 0.048 |
2.801 ± 0.037 | 2.444 ± 0.026 |
4.327 ± 0.049 | 6.053 ± 0.049 |
4.857 ± 0.047 | 6.83 ± 0.051 |
0.718 ± 0.018 | 3.676 ± 0.039 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
